This Jupyter notebook can be downloaded from paper_validation_example.ipynb, or viewed as a python script at paper_validation_example.py.
Validation Example for PINT paper
A comparison between PINT result and Tempo/Tempo2 result. This example is presented in the PINT paper to validate that PINT is able to process the PSR J1600-3053 NANOGrav 11-year data set, which uses DD binary model, and get a comparable result with TEMPO/TEMPO2. For more discussion see: https://arxiv.org/abs/2012.00074
Method of this comparison:
Requirement: * Data set: PRS J1600-3053 NANOGrav 11-year data * One copy of PRS J1600-3053 NANOGrav 11-year data is included in the PINT source code docs/examples/J1600-3053_NANOGrav_11yv1.gls.par and docs/examples/J1600-3053_NANOGrav_11yv1.tim, which is the default data path in this notebook. Note, this requires the user to download the PINT source code from github. * The offical NANOGrav 11-year data can be downloaded at: https://data.nanograv.org/. The data path should be changed
to the data location. * PINT version: 0.8.0 or higher. * TEMPO and its python utils tempo_utils. * TEMPO version for current comparison: 13.101 (2020-11-04 c5fbddf) * TEMPO2 and its python utils tempo2_utils. * TEMPO2 version for current comparison: 2019.01.1 * TEMPO_utils and TEMPO2_utils are packaged together and can be downloaded from https://github.com/demorest/tempo_utils. * TEMPO2 general2 plugins.
[1]:
import pint
import sys
from pint import toa
from pint import models
from pint.fitter import GLSFitter
import os
import matplotlib.pyplot as plt
import astropy.units as u
import tempo2_utils as t2u
import tempo_utils
import tempo2_utils
import numpy as np
from astropy.table import Table
from astropy.io import ascii
import subprocess
import tempfile
from pint import ls
import astropy.constants as ct
from pint.solar_system_ephemerides import objPosVel_wrt_SSB
from astropy.time import Time
Print the PINT and TEMPO/TEMPO2 version
[2]:
print("PINT version: ", pint.__version__)
tempo_v = subprocess.check_output(["tempo", "-v"])
print("TEMPO version: ", tempo_v.decode("utf-8"))
#Not sure why tempo2_v = subprocess.check_output(["tempo2", "-v"]) does not work.
process = subprocess.Popen(['tempo2', '-v'], stdout=subprocess.PIPE)
tempo2_v = process.communicate()[0]
print("TEMPO2 version: ", tempo2_v.decode("utf-8"))
PINT version: 0.8+68.g6c072c27
TEMPO version: Tempo v 13.101 (2020-11-04 c5fbddf)
TEMPO2 version: 2019.01.1
Redefine the Tempo2_util function for larger number of observations
[3]:
_nobs = 30000
def newpar2(parfile,timfile):
"""
Run tempo2, return new parfile (as list of lines). input parfile
can be either lines or a filename.
"""
orig_dir = os.getcwd()
try:
temp_dir = tempfile.mkdtemp(prefix="tempo2")
try:
lines = open(parfile,'r').readlines()
except:
lines = parfile
open("%s/pulsar.par" % temp_dir, 'w').writelines(lines)
timpath = os.path.abspath(timfile)
os.chdir(temp_dir)
cmd = "tempo2 -nobs %d -newpar -f pulsar.par %s -norescale" % (_nobs, timpath)
os.system(cmd + " > /dev/null")
outparlines = open('new.par').readlines()
finally:
os.chdir(orig_dir)
os.system("rm -rf %s" % temp_dir)
for l in outparlines:
if l.startswith('TRES'): rms = float(l.split()[1])
elif l.startswith('CHI2R'): (foo, chi2r, ndof) = l.split()
return float(chi2r)*float(ndof), int(ndof), rms, outparlines
Set up date file path for PSR J1600-3053.
Note
This path only works when PINT is installed from source code, which has docs and example directories.
[4]:
psr = "J1600-3053"
par_file = os.path.join('../examples', psr + "_NANOGrav_11yv1.gls.par")
tim_file = os.path.join('../examples', psr + "_NANOGrav_11yv1.tim")
PINT run
Load TOAs to PINT
[5]:
t = toa.get_TOAs(tim_file, ephem="DE436", bipm_version="BIPM2015")
INFO: Applying clock corrections (include_gps = True, include_bipm = True) [pint.toa]
INFO: Observatory gbt, loading clock file
/home/luo/.local/lib/python3.6/site-packages/pint/datafiles/time.dat [pint.observatory.topo_obs]
INFO: Applying observatory clock corrections. [pint.observatory.topo_obs]
INFO: Applying GPS to UTC clock correction (~few nanoseconds) [pint.observatory.topo_obs]
INFO: Observatory gbt, loading GPS clock file
/home/luo/.local/lib/python3.6/site-packages/pint/datafiles/gps2utc.clk [pint.observatory.topo_obs]
INFO: Applying TT(TAI) to TT(BIPM2015) clock correction (~27 us) [pint.observatory.topo_obs]
INFO: Observatory gbt, loading BIPM clock file
/home/luo/.local/lib/python3.6/site-packages/pint/datafiles/tai2tt_bipm2015.clk [pint.observatory.topo_obs]
INFO: Computing TDB columns. [pint.toa]
INFO: Using EPHEM = DE436 for TDB calculation. [pint.toa]
INFO: Computing PosVels of observatories and Earth, using DE436 [pint.toa]
INFO: Set solar system ephemeris to link:
https://data.nanograv.org/static/data/ephem/de436.bsp [pint.solar_system_ephemerides]
[6]:
print("There are {} TOAs in the dataset.".format(t.ntoas))
There are 12433 TOAs in the dataset.
Load timing model from .par file
Since PINT only uses the IAU 2000 version of precession-nutation model but NANOGrav 11-year data uses old precession-nutation model, You will see a UserWarning: PINT only supports 'T2CMETHOD IAU2000B'.
[7]:
m = models.get_model(par_file)
INFO: Parameter A1DOT's value will be scaled by 1e-12 [pint.models.parameter]
INFO: Parameter A1DOT's value will be scaled by 1e-12 [pint.models.parameter]
/home/luo/.local/lib/python3.6/site-packages/pint/models/timing_model.py:304: UserWarning: PINT only supports 'T2CMETHOD IAU2000B'
warn("PINT only supports 'T2CMETHOD IAU2000B'")
Make the General Least Square fitter
[8]:
f = GLSFitter(model=m, toas=t)
Fit TOAs for 9 iterations.
The expected chi2 value should be close to TEMPO and TEMPO2, but not the same.
[9]:
chi2 = f.fit_toas(9)
print("Postfit Chi2: ", chi2)
print("Degree of freedom: ", f.resids.dof)
/home/luo/.local/lib/python3.6/site-packages/pint/models/timing_model.py:304: UserWarning: PINT only supports 'T2CMETHOD IAU2000B'
warn("PINT only supports 'T2CMETHOD IAU2000B'")
/home/luo/.local/lib/python3.6/site-packages/pint/models/timing_model.py:304: UserWarning: PINT only supports 'T2CMETHOD IAU2000B'
warn("PINT only supports 'T2CMETHOD IAU2000B'")
/home/luo/.local/lib/python3.6/site-packages/pint/models/timing_model.py:304: UserWarning: PINT only supports 'T2CMETHOD IAU2000B'
warn("PINT only supports 'T2CMETHOD IAU2000B'")
/home/luo/.local/lib/python3.6/site-packages/pint/models/timing_model.py:304: UserWarning: PINT only supports 'T2CMETHOD IAU2000B'
warn("PINT only supports 'T2CMETHOD IAU2000B'")
/home/luo/.local/lib/python3.6/site-packages/pint/models/timing_model.py:304: UserWarning: PINT only supports 'T2CMETHOD IAU2000B'
warn("PINT only supports 'T2CMETHOD IAU2000B'")
/home/luo/.local/lib/python3.6/site-packages/pint/models/timing_model.py:304: UserWarning: PINT only supports 'T2CMETHOD IAU2000B'
warn("PINT only supports 'T2CMETHOD IAU2000B'")
/home/luo/.local/lib/python3.6/site-packages/pint/models/timing_model.py:304: UserWarning: PINT only supports 'T2CMETHOD IAU2000B'
warn("PINT only supports 'T2CMETHOD IAU2000B'")
/home/luo/.local/lib/python3.6/site-packages/pint/models/timing_model.py:304: UserWarning: PINT only supports 'T2CMETHOD IAU2000B'
warn("PINT only supports 'T2CMETHOD IAU2000B'")
/home/luo/.local/lib/python3.6/site-packages/pint/models/timing_model.py:304: UserWarning: PINT only supports 'T2CMETHOD IAU2000B'
warn("PINT only supports 'T2CMETHOD IAU2000B'")
/home/luo/.local/lib/python3.6/site-packages/pint/models/timing_model.py:304: UserWarning: PINT only supports 'T2CMETHOD IAU2000B'
warn("PINT only supports 'T2CMETHOD IAU2000B'")
Postfit Chi2: 12368.094375740437515
Degree of freedom: 12307
The weighted RMS value for pre-fit and post-fit residuals
[10]:
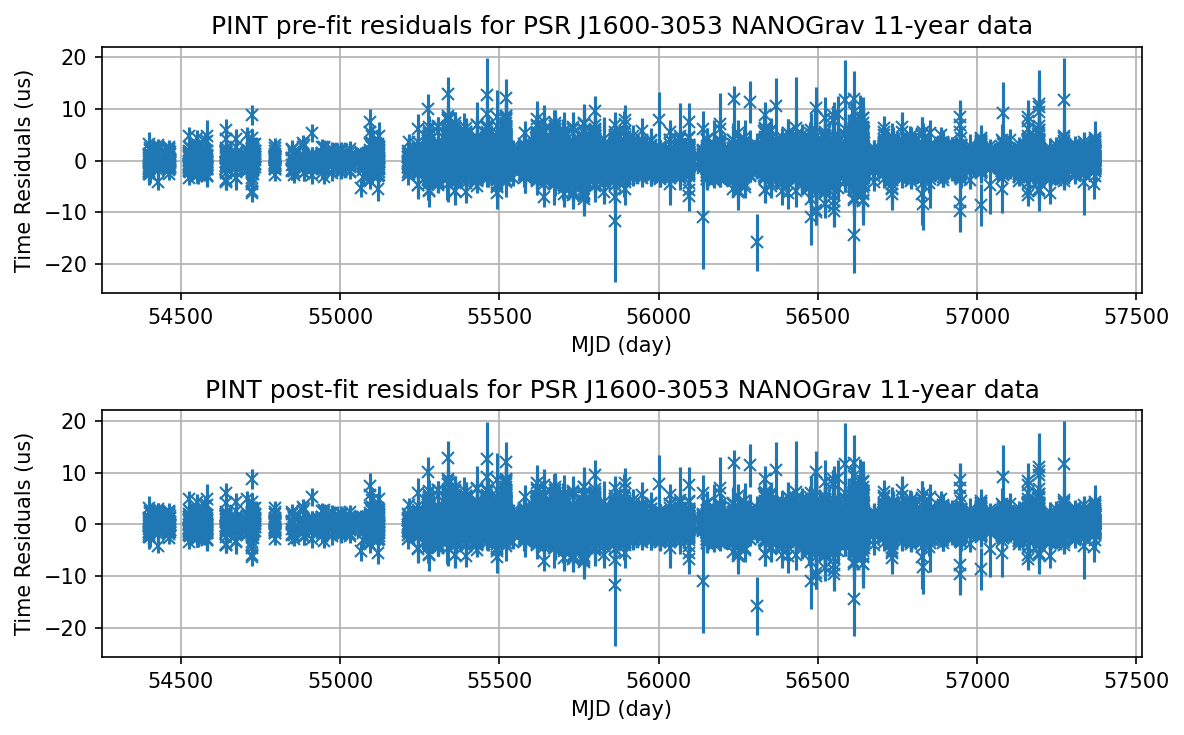
print("Pre-fit residual weighted RMS:", f.resids_init.rms_weighted())
print("Post-fit residual weighted RMS:", f.resids.rms_weighted())
Pre-fit residual weighted RMS: 0.944170684867224 us
Post-fit residual weighted RMS: 0.9441138383219785 us
Plot the pre-fit and post-fit residuals
[11]:
pint_prefit = f.resids_init.time_resids.to_value(u.us)
pint_postfit = f.resids.time_resids.to_value(u.us)
plt.figure(figsize=(8,5), dpi=150)
plt.subplot(2, 1, 1)
plt.errorbar(t.get_mjds().to_value(u.day), f.resids_init.time_resids.to_value(u.us),
yerr=t.get_errors().to_value(u.us), fmt='x')
plt.xlabel('MJD (day)')
plt.ylabel('Time Residuals (us)')
plt.title('PINT pre-fit residuals for PSR J1600-3053 NANOGrav 11-year data')
plt.grid(True)
plt.subplot(2, 1, 2)
plt.errorbar(t.get_mjds().to_value(u.day), f.resids.time_resids.to_value(u.us),
yerr=t.get_errors().to_value(u.us), fmt='x')
plt.xlabel('MJD (day)')
plt.ylabel('Time Residuals (us)')
plt.title('PINT post-fit residuals for PSR J1600-3053 NANOGrav 11-year data')
plt.grid(True)
plt.tight_layout()
plt.savefig("J1600_PINT")
TEMPO run
Use tempo_utils to analysis the same data set.
[12]:
tempo_toa = tempo_utils.read_toa_file(tim_file)
tempo_chi2, ndof, rms_t, tempo_par = tempo_utils.run_tempo(tempo_toa ,par_file, get_output_par=True,
gls=True)
[13]:
print("TEMPO postfit chi2: ", tempo_chi2)
print("TEMPO postfit weighted rms: ", rms_t)
TEMPO postfit chi2: 12368.46
TEMPO postfit weighted rms: 0.944
Write the TEMPO postfit model to a new .par file, for comparison later
[14]:
# Write out the post fit tempo parfile.
tempo_parfile = open(psr + '_tempo.par', 'w')
for line in tempo_par:
tempo_parfile.write(line)
tempo_parfile.close()
Get the TEMPO residuals
[15]:
tempo_prefit = tempo_toa.get_prefit()
tempo_postfit = tempo_toa.get_resids()
mjds = tempo_toa.get_mjd()
freqs = tempo_toa.get_freq()
errs = tempo_toa.get_resid_err()
Plot the PINT - TEMPO residual difference.
[16]:
tp_diff_pre = (pint_prefit - tempo_prefit) * u.us
tp_diff_post = (pint_postfit - tempo_postfit) * u.us
[17]:
plt.figure(figsize=(8,5), dpi=150)
plt.subplot(2, 1, 1)
plt.plot(mjds, (tp_diff_pre - tp_diff_pre.mean()).to_value(u.ns), '+')
plt.xlabel('MJD (day)')
plt.ylabel('Time Residuals (ns)')
plt.title('PSR J1600-3053 prefit residual differences between PINT and TEMPO')
plt.grid(True)
plt.subplot(2, 1, 2)
plt.plot(mjds, (tp_diff_post - tp_diff_post.mean()).to_value(u.ns), '+')
plt.xlabel('MJD (day)')
plt.ylabel('Time Residuals (ns)')
plt.title('PSR J1600-3053 postfit residual differences between PINT and TEMPO')
plt.grid(True)
plt.tight_layout()
plt.savefig("J1600_PINT_tempo.eps")
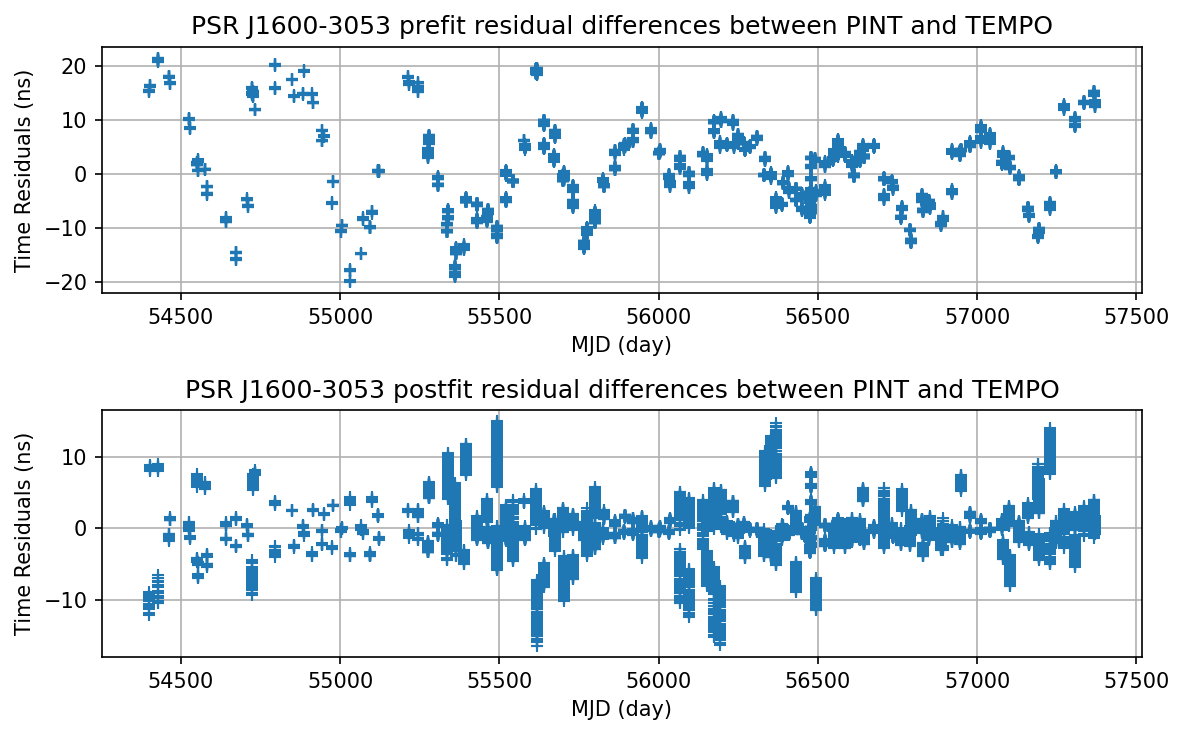
The PINT-TEMPO pre-fit residual discrepancy is due to the different precession-nutation model in the two packages. * TEMPO: IAU6501976 precession and IAU 1980 nutation. * PINT: IAU2000B precession-nutation.
Compare the parameter between TEMPO and PINT
Reported quantities
TEMPO value
TEMPO uncertainty
Parameter units
TEMPO parameter value - PINT parameter value
TEMPO/PINT parameter absolute difference divided by TEMPO uncertainty
PINT uncertainty divided by TEMPO uncertainty, if TEMPO provides the uncertainty value
[18]:
# Create the parameter compare table
tv = []
tu = []
tv_pv = []
tv_pv_tc = []
tc_pc = []
units = []
names = []
no_t_unc = []
tempo_new_model = models.get_model(psr + '_tempo.par')
for param in tempo_new_model.params:
t_par = getattr(tempo_new_model, param)
pint_par = getattr(f.model, param)
tempoq = t_par.quantity
pintq = pint_par.quantity
try:
diffq = tempoq - pintq
if t_par.uncertainty_value != 0.0:
diff_tcq = np.abs(diffq) / t_par.uncertainty
uvsu = pint_par.uncertainty / t_par.uncertainty
no_t_unc.append(False)
else:
diff_tcq = np.abs(diffq) / pint_par.uncertainty
uvsu = t_par.uncertainty
no_t_unc.append(True)
except TypeError:
continue
uvsu = pint_par.uncertainty / t_par.uncertainty
tv.append(tempoq.value)
tu.append(t_par.uncertainty.value)
tv_pv.append(diffq.value)
tv_pv_tc.append(diff_tcq.value)
tc_pc.append(uvsu)
units.append(t_par.units)
names.append(param)
compare_table = Table((names, tv, tu, units, tv_pv, tv_pv_tc, tc_pc, no_t_unc), names = ('name', 'Tempo Value', 'Tempo uncertainty', 'units',
'Tempo_V-PINT_V',
'Tempo_PINT_diff/unct',
'PINT_unct/Tempo_unct',
'no_t_unc'))
compare_table.sort('Tempo_PINT_diff/unct')
compare_table = compare_table[::-1]
compare_table.write('parameter_compare.t.html', format='html', overwrite=True)
INFO: Parameter A1DOT's value will be scaled by 1e-12 [pint.models.parameter]
INFO: Parameter A1DOT's value will be scaled by 1e-12 [pint.models.parameter]
/home/luo/.local/lib/python3.6/site-packages/pint/models/timing_model.py:304: UserWarning: PINT only supports 'T2CMETHOD IAU2000B'
warn("PINT only supports 'T2CMETHOD IAU2000B'")
Print the parameter difference in a table.
The table is sorted by relative difference in descending order.
[19]:
compare_table
[19]:
| name | Tempo Value | Tempo uncertainty | units | Tempo_V-PINT_V | Tempo_PINT_diff/unct | PINT_unct/Tempo_unct | no_t_unc |
|---|---|---|---|---|---|---|---|
| str8 | str48 | float128 | object | float128 | float128 | float128 | bool |
| ELONG | 244.347677844079 | 5.9573e-09 | deg | -5.923646156924533557e-10 | 0.09943508228433239147 | 0.99997665908491340815 | False |
| ELAT | -10.0718390253651 | 3.36103e-08 | deg | -3.1908024911500576515e-09 | 0.09493525767845147623 | 1.0000723294528560605 | False |
| PMELONG | 0.4626 | 0.010399999999999999523 | mas / yr | 0.00071186955533908413685 | 0.06844899570568116487 | 1.0031591658256824307 | False |
| F0 | 277.9377112429746148 | 5.186e-13 | Hz | -1.471045507628332416e-14 | 0.028365705893334601157 | 1.0000737060332136081 | False |
| PX | 0.504 | 0.07349999999999999589 | mas | -0.0020714040496805363745 | 0.028182368022864442286 | 0.99982583641940803165 | False |
| ECC | 0.0001737294 | 8.9000000000000002855e-09 | -2.385671795204248602e-10 | 0.026805301069710657513 | 1.0022775158955208319 | False | |
| DMX_0010 | 0.00066927561 | 0.00020051850499999999489 | pc / cm3 | -5.0888210248638612865e-06 | 0.025378311218028786617 | 0.9999978605214362437 | False |
| DMX_0001 | 0.0016432056 | 0.00022434462499999998828 | pc / cm3 | -5.3330253040537178855e-06 | 0.023771576003007506561 | 1.0000068595527518145 | False |
| DMX_0002 | 0.00136024872 | 0.00020941304000000001188 | pc / cm3 | -4.909940340062750319e-06 | 0.023446201535791421494 | 1.0000106543543163529 | False |
| OM | 181.84956816578 | 0.01296546975 | deg | -0.00026376139872756609872 | 0.020343373885667821539 | 0.9909564544661490358 | False |
| ... | ... | ... | ... | ... | ... | ... | ... |
| DMX_0045 | 3.64190777e-05 | 0.00020164094999999999935 | pc / cm3 | -1.07417857443502110307e-07 | 0.00053271846538861333895 | 1.00000068370106443 | False |
| DMX_0071 | -0.000176912603 | 0.00019118353399999999634 | pc / cm3 | -9.7209239643334917694e-08 | 0.0005084603135505116932 | 1.0000046197730818598 | False |
| DMX_0075 | 2.00017094e-06 | 0.00019663653799999999744 | pc / cm3 | -9.568337454155456868e-08 | 0.00048660017876003580943 | 0.9999419901870312266 | False |
| DMX_0094 | 0.000929849121 | 0.00019402737299999999105 | pc / cm3 | 4.607091936333768123e-08 | 0.0002374454627251881909 | 0.99999408921119359306 | False |
| DMX_0073 | -0.000156953835 | 0.00019724444300000000259 | pc / cm3 | 4.614141422537424743e-08 | 0.00023393010988590560973 | 1.0000039448402757714 | False |
| DMX_0017 | 0.000178762757 | 0.00021197504699999999088 | pc / cm3 | -3.7177033512374732874e-08 | 0.00017538400881861692499 | 0.99998893629003005046 | False |
| DMX_0067 | -0.000377967984 | 0.00019749766400000001308 | pc / cm3 | -2.135996082108402791e-08 | 0.00010815297957693325822 | 0.9999769537566483013 | False |
| DMX_0043 | -0.000494848648 | 0.0001997188189999999947 | pc / cm3 | 1.8844699835535508314e-08 | 9.43561549677274415e-05 | 0.99998484576198187757 | False |
| DMX_0083 | 8.70047706e-06 | 0.00020486178099999999887 | pc / cm3 | 1.8072403983782853805e-08 | 8.8217547927023319e-05 | 1.0000060515174629128 | False |
| DMX_0069 | -0.000251368356 | 0.00019942850700000000919 | pc / cm3 | 1.6777396572118397772e-08 | 8.412737388701604967e-05 | 1.0000028589159370984 | False |
If one wants the Latex output please use the cell below.
[20]:
#ascii.write(compare_table, sys.stdout, Writer = ascii.Latex,
# latexdict = {'tabletype': 'table*'})
Check out the maximum DMX difference
[21]:
max_dmx = 0
max_dmx_index = 0
for ii, row in enumerate(compare_table):
if row['name'].startswith('DMX_'):
if row['Tempo_PINT_diff/unct'] > max_dmx:
max_dmx = row['Tempo_PINT_diff/unct']
max_dmx_index = ii
dmx_max = compare_table[max_dmx_index]['name']
compare_table[max_dmx_index]
[21]:
| name | Tempo Value | Tempo uncertainty | units | Tempo_V-PINT_V | Tempo_PINT_diff/unct | PINT_unct/Tempo_unct | no_t_unc |
|---|---|---|---|---|---|---|---|
| str8 | str48 | float128 | object | float128 | float128 | float128 | bool |
| DMX_0010 | 0.00066927561 | 0.00020051850499999999489 | pc / cm3 | -5.0888210248638612865e-06 | 0.025378311218028786617 | 0.9999978605214362437 | False |
Output the table in the paper
[22]:
paper_params = ['F0', 'F1', 'FD1', 'FD2', 'JUMP1', 'PX',
'ELONG', 'ELAT', 'PMELONG', 'PMELAT', 'PB',
'A1', 'A1DOT', 'ECC', 'T0', 'OM', 'OMDOT', 'M2',
'SINI', dmx_max]
# Get the table index of the parameters above
paper_param_index = []
for pp in paper_params:
# We assume the parameter name are unique in the table
idx = np.where(compare_table['name'] == pp)[0][0]
paper_param_index.append(idx)
paper_param_index = np.array(paper_param_index)
compare_table[paper_param_index]
[22]:
| name | Tempo Value | Tempo uncertainty | units | Tempo_V-PINT_V | Tempo_PINT_diff/unct | PINT_unct/Tempo_unct | no_t_unc |
|---|---|---|---|---|---|---|---|
| str8 | str48 | float128 | object | float128 | float128 | float128 | bool |
| F0 | 277.9377112429746148 | 5.186e-13 | Hz | -1.471045507628332416e-14 | 0.028365705893334601157 | 1.0000737060332136081 | False |
| F1 | -7.338737472765e-16 | 4.619148184227e-21 | Hz / s | 6.3620434143467849095e-23 | 0.013773196183814252269 | 1.0001125817340140925 | False |
| FD1 | 3.98314325e-05 | 1.6566479199999999207e-06 | s | -2.5460168459788480762e-09 | 0.0015368484849688811896 | 0.9999972210804803918 | False |
| FD2 | -1.47296057e-05 | 1.1922595999999999884e-06 | s | 1.3701818969060749554e-09 | 0.0011492311715553180356 | 0.99999858894148907495 | False |
| JUMP1 | -8.789e-06 | 1.2999999999999999941e-07 | s | -4.6499257614788852208e-10 | 0.0035768659703683731467 | 1.0037094619649047367 | False |
| PX | 0.504 | 0.07349999999999999589 | mas | -0.0020714040496805363745 | 0.028182368022864442286 | 0.99982583641940803165 | False |
| ELONG | 244.347677844079 | 5.9573e-09 | deg | -5.923646156924533557e-10 | 0.09943508228433239147 | 0.99997665908491340815 | False |
| ELAT | -10.0718390253651 | 3.36103e-08 | deg | -3.1908024911500576515e-09 | 0.09493525767845147623 | 1.0000723294528560605 | False |
| PMELONG | 0.4626 | 0.010399999999999999523 | mas / yr | 0.00071186955533908413685 | 0.06844899570568116487 | 1.0031591658256824307 | False |
| PMELAT | -7.1555 | 0.058200000000000001732 | mas / yr | -0.00050484895274838237356 | 0.008674380631415503848 | 0.9992173822396303029 | False |
| PB | 14.34846572550302 | 2.1222661e-06 | d | -3.4570101856666590745e-08 | 0.016289240004666045763 | 1.0000725690593949082 | False |
| A1 | 8.801653122 | 8.1100000000000004906e-07 | ls | 1.4913034362962207524e-08 | 0.018388451742246864767 | 0.9844174919313783967 | False |
| A1DOT | -4e-15 | 6.260000000000000155e-16 | ls / s | 8.912568233346704436e-18 | 0.014237329446240742231 | 0.99986821703571171494 | False |
| ECC | 0.0001737294 | 8.9000000000000002855e-09 | -2.385671795204248602e-10 | 0.026805301069710657513 | 1.0022775158955208319 | False | |
| T0 | 55878.2618980451000000 | 0.0005167676 | d | -1.0512794592798524462e-05 | 0.020343370197354718952 | 0.9909423736001105048 | False |
| OM | 181.84956816578 | 0.01296546975 | deg | -0.00026376139872756609872 | 0.020343373885667821539 | 0.9909564544661490358 | False |
| OMDOT | 0.0052209 | 0.0013554 | deg / yr | -2.2108721249702442383e-05 | 0.016311584218461297317 | 1.0000993017977168461 | False |
| M2 | 0.271894 | 0.089418999999999998485 | solMass | -0.0016411381726809115555 | 0.018353349653663222213 | 0.97866231479642251667 | False |
| SINI | 0.906285 | 0.03399300000000000238 | 0.00054355868381605887407 | 0.015990312235344302655 | 0.9838880245552520387 | False | |
| DMX_0010 | 0.00066927561 | 0.00020051850499999999489 | pc / cm3 | -5.0888210248638612865e-06 | 0.025378311218028786617 | 0.9999978605214362437 | False |
TEMPO2 run
Before TEMPO2 run, the .par file has to be modified for a more accurate TEMPO2 vs PINT comparison. We save the modified .par file in a file named “[PSR name]_tempo2.par”. In this case, “J1600-3053_tempo2.par”
Modified parameters in the .par file:
ECL IERS2010 —-> ECL IERS 2003 (In this version of TEMPO2, the IERS 2003 Obliquity angle is hardcoded in its code. To match TEMPO2’s default value, we change the ECL to IERS 2003 in the
.parfile )T2CMETHOD TEMPO —-> # T2CMETHOD TEMPO (TEMPO2 supports both IAU 2000 precession-nutation model and old TEMPO-style model. To make TEMPO2 ues its default precession and nutation model, IAU 2000, this line in the
.parfile has to be commmented out.)
Note, this modified
.parfile is provided in thedocs/examplesdirectory. If PINT is not installed from source code, one have to modify the.parfile from the NANOGrav 11-year data.
[23]:
tempo2_par = os.path.join('../examples',"J1600-3053_tempo2.par")
PINT refit using the modified tempo2-style parfile
[24]:
m_t2 = models.get_model(tempo2_par)
INFO: Parameter A1DOT's value will be scaled by 1e-12 [pint.models.parameter]
INFO: Parameter A1DOT's value will be scaled by 1e-12 [pint.models.parameter]
[25]:
f_t2 = GLSFitter(toas=t, model=m_t2)
f_t2.fit_toas()
[25]:
12368.092265853045861
Tempo2 fit
[26]:
tempo2_chi2, ndof, rms_t2, tempo2_new_par = newpar2(tempo2_par, tim_file)
print("TEMPO2 chi2: ", tempo2_chi2)
print("TEMPO2 rms: ", rms_t2)
TEMPO2 chi2: 12265.156200000001
TEMPO2 rms: 0.944
Get TEMPO2 residuals, toa value, observing frequencies, and data error
[27]:
tempo2_result = t2u.general2(tempo2_par, tim_file, ['sat', 'pre', 'post', 'freq', 'err'])
# TEMPO2's residual unit is second
tp2_diff_pre = f_t2.resids_init.time_resids - tempo2_result['pre'] * u.s
tp2_diff_post = f_t2.resids.time_resids - tempo2_result['post'] * u.s
Plot the TEMPO2 - PINT residual difference
[28]:
plt.figure(figsize=(8,5), dpi=150)
plt.subplot(2, 1, 1)
plt.plot(mjds, (tp2_diff_pre - tp2_diff_pre.mean()).to_value(u.ns), '+')
plt.xlabel('MJD (day)')
plt.ylabel('Time Residuals (ns)')
plt.title('PSR J1600-3053 prefit residual differences between PINT and TEMPO2')
plt.grid(True)
plt.subplot(2, 1, 2)
plt.plot(mjds, (tp2_diff_post - tp2_diff_post.mean()).to_value(u.ns), '+')
plt.xlabel('MJD (day)')
plt.ylabel('Time Residuals (ns)')
plt.title('PSR J1600-3053 postfit residual differences between PINT and TEMPO2')
plt.grid(True)
plt.tight_layout()
plt.savefig("J1600_PINT_tempo2")
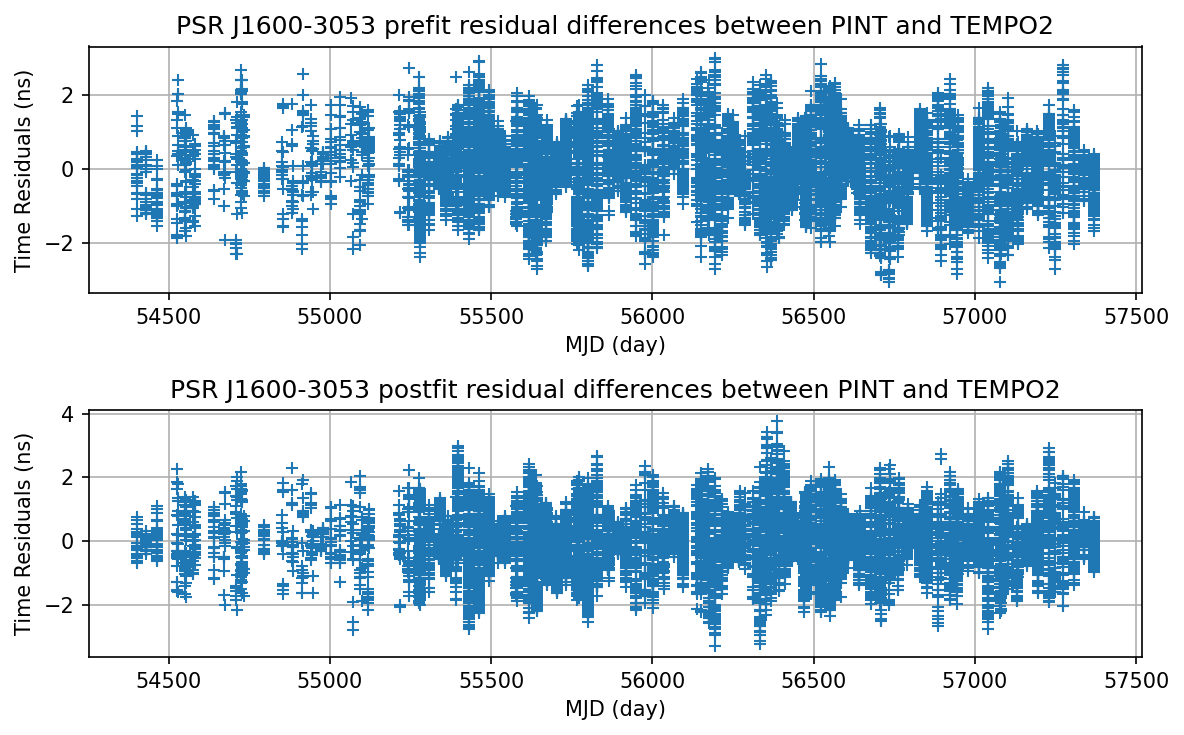
In this comparison, PINT and TEMPO2’s results, both pre-fit and post-fit, agree with each other within the level of 5 ns.
Write out the TEMPO2 postfit parameter to a new file
Note, since the ECL parameter is hard coded in tempo2, we will have to add it manually
[29]:
# Write out the post fit tempo parfile.
tempo2_parfile = open(psr + '_new_tempo2.2.par', 'w')
for line in tempo2_new_par:
tempo2_parfile.write(line)
tempo2_parfile.write("ECL IERS2003")
tempo2_parfile.close()
Compare the parameter between TEMPO2 and PINT
Reported quantities
TEMPO2 value
TEMPO2 uncertainty
Parameter units
TEMPO2 parameter value - PINT parameter value
TEMPO2/PINT parameter absolute difference divided by TEMPO2 uncertainty
PINT uncertainty divided by TEMPO2 uncertainty, if TEMPO2 provides the uncertainty value
[30]:
# Create the parameter compare table
tv = []
t2_unc = []
tv_pv = []
tv_pv_tc = []
tc_pc = []
units = []
names = []
no_t2_unc = []
tempo2_new_model = models.get_model(psr + '_new_tempo2.2.par')
for param in tempo2_new_model.params:
t2_par = getattr(tempo2_new_model, param)
pint2_par = getattr(f_t2.model, param)
tempo2q = t2_par.quantity
pint2q = pint2_par.quantity
try:
diff2q = tempo2q - pint2q
if t2_par.uncertainty_value != 0.0:
diff_tcq = np.abs(diff2q) / t2_par.uncertainty
uvsu = pint2_par.uncertainty / t2_par.uncertainty
no_t2_unc.append(False)
else:
diff_tcq = np.abs(diff2q) / pint2_par.uncertainty
uvsu = t2_par.uncertainty
no_t2_unc.append(True)
except TypeError:
continue
uvsu = pint2_par.uncertainty / t2_par.uncertainty
tv.append(tempo2q.value)
t2_unc.append(t2_par.uncertainty.value)
tv_pv.append(diff2q.value)
tv_pv_tc.append(diff_tcq.value)
tc_pc.append(uvsu)
units.append(t2_par.units)
names.append(param)
compare_table2 = Table((names, tv, t2_unc,units, tv_pv, tv_pv_tc, tc_pc, no_t2_unc), names = ('name', 'Tempo2 Value', 'T2 unc','units',
'Tempo2_V-PINT_V',
'Tempo2_PINT_diff/unct',
'PINT_unct/Tempo2_unct',
'no_t_unc'))
compare_table2.sort('Tempo2_PINT_diff/unct')
compare_table2 = compare_table2[::-1]
compare_table2.write('parameter_compare.t2.html', format='html', overwrite=True)
WARNING: EPHVER 5 does nothing in PINT [pint.models.timing_model]
WARNING: Unrecognized parfile line 'NE_SW 0' [pint.models.timing_model]
WARNING: Unrecognized parfile line 'NE_SW2 0.000' [pint.models.timing_model]
/home/luo/.local/lib/python3.6/site-packages/astropy/units/quantity.py:477: RuntimeWarning: divide by zero encountered in true_divide
result = super().__array_ufunc__(function, method, *arrays, **kwargs)
Print the parameter difference in a table.
The table is sorted by relative difference in descending order.
[31]:
compare_table2
[31]:
| name | Tempo2 Value | T2 unc | units | Tempo2_V-PINT_V | Tempo2_PINT_diff/unct | PINT_unct/Tempo2_unct | no_t_unc |
|---|---|---|---|---|---|---|---|
| str8 | str48 | float128 | object | float128 | float128 | float128 | bool |
| ECC | 0.00017372966157521168 | 8.922286680669999241e-09 | 4.158524756102052744e-11 | 0.0046608284455950449096 | 1.0000400945849889922 | False | |
| DMX_0098 | 0.0013394613122489417 | 0.00019579968831114546654 | pc / cm3 | -5.203720844662671624e-07 | 0.002657675755026450183 | 0.999999270898346615 | False |
| DMX_0070 | -0.00023747963906517973 | 0.00019767137320477682749 | pc / cm3 | -4.67328571424551701e-07 | 0.0023641691958118014015 | 1.0000006149827960211 | False |
| DMX_0097 | 0.0013928330661987446 | 0.00019620100461426303326 | pc / cm3 | -4.3986846819175744183e-07 | 0.002241927706010230642 | 0.9999998585691036723 | False |
| DMX_0055 | -0.0005307704904403621 | 0.00019675128861832102923 | pc / cm3 | -3.9826563099699431592e-07 | 0.0020242085009648507626 | 1.0000000155754873443 | False |
| DMX_0063 | -0.00048410571072825574 | 0.00019894769104906708185 | pc / cm3 | -3.8863084052005031355e-07 | 0.001953432273934765668 | 1.0000001764728303488 | False |
| DMX_0079 | 0.00018976795294000216 | 0.00019490725481464179483 | pc / cm3 | -3.6899448284041791446e-07 | 0.001893179826432496481 | 1.000000186999052243 | False |
| DMX_0010 | 0.00067403356955979 | 0.00020051850482404336064 | pc / cm3 | -3.7725252445390284467e-07 | 0.0018813850860545018075 | 0.99999987947730495375 | False |
| F1 | -7.3387383041227678664e-16 | 4.619148404392432094e-21 | Hz / s | -8.1916189706268323405e-24 | 0.0017734045874857090789 | 0.9999982063295332385 | False |
| DMX_0086 | 0.00029525346690830644 | 0.0001961188165133768578 | pc / cm3 | -3.4520664969226941277e-07 | 0.001760191376989691343 | 1.0000003907045813545 | False |
| ... | ... | ... | ... | ... | ... | ... | ... |
| DMX_0092 | 0.0013207295138539894 | 0.00019585216459019454951 | pc / cm3 | -3.4341162511537445812e-08 | 0.00017534226687457687001 | 1.0000000634867269866 | False |
| DMX_0058 | -0.0005377581468744793 | 0.00019927530538964258904 | pc / cm3 | -3.1767895307833332597e-08 | 0.00015941712017812561131 | 1.0000003635542755731 | False |
| DMX_0017 | 0.0001789867729265184 | 0.00021197504981685315979 | pc / cm3 | 2.854267636918103937e-08 | 0.000134651112920327015 | 0.9999995235765041235 | False |
| DMX_0089 | 0.0007495614446295846 | 0.00021586616414944812654 | pc / cm3 | 2.4131246270523223907e-08 | 0.000111787997742048708454 | 1.0000000332176062212 | False |
| DMX_0027 | -0.00018288082535181414 | 0.00019391445756469536201 | pc / cm3 | -1.7893991355739631913e-08 | 9.227775783437751106e-05 | 1.000000166662465162 | False |
| DMX_0040 | -0.0005242449385393532 | 0.00020212647115737782458 | pc / cm3 | -1.6804078603323106822e-08 | 8.313645663085500665e-05 | 0.99999999749641554914 | False |
| DMX_0032 | -6.265675469663684e-05 | 0.00019561483985536690729 | pc / cm3 | 1.5367171419820358814e-08 | 7.855831097059144273e-05 | 1.0000001097328992117 | False |
| DMX_0001 | 0.0016484372168232325 | 0.00022434462780433157077 | pc / cm3 | -1.4668063582322365956e-08 | 6.538183564224023551e-05 | 1.0000004801524928766 | False |
| DMX_0083 | 8.544780315309648e-06 | 0.00020486177918444288125 | pc / cm3 | -9.68929340697572577e-09 | 4.729673561143965028e-05 | 1.0000001554708373153 | False |
| DMX_0044 | -0.0003390023662491028 | 0.00021062295971768858391 | pc / cm3 | -4.0461326309814554802e-09 | 1.9210311337399994e-05 | 1.000000188731715367 | False |
If one wants to get the latex version, please use the line below.
[32]:
#ascii.write(compare_table2, sys.stdout, Writer = ascii.Latex,
# latexdict = {'tabletype': 'table*'})
Check out the maximum DMX difference
[33]:
max_dmx = 0
max_dmx_index = 0
for ii, row in enumerate(compare_table2):
if row['name'].startswith('DMX_'):
if row['Tempo2_PINT_diff/unct'] > max_dmx:
max_dmx = row['Tempo2_PINT_diff/unct']
max_dmx_index = ii
dmx_max2 = compare_table2[max_dmx_index]['name']
compare_table2[max_dmx_index]
[33]:
| name | Tempo2 Value | T2 unc | units | Tempo2_V-PINT_V | Tempo2_PINT_diff/unct | PINT_unct/Tempo2_unct | no_t_unc |
|---|---|---|---|---|---|---|---|
| str8 | str48 | float128 | object | float128 | float128 | float128 | bool |
| DMX_0098 | 0.0013394613122489417 | 0.00019579968831114546654 | pc / cm3 | -5.203720844662671624e-07 | 0.002657675755026450183 | 0.999999270898346615 | False |
Output the table in the paper
[34]:
paper_params = ['F0', 'F1', 'FD1', 'FD2', 'JUMP1', 'PX',
'ELONG', 'ELAT', 'PMELONG', 'PMELAT', 'PB',
'A1', 'A1DOT', 'ECC', 'T0', 'OM', 'OMDOT', 'M2',
'SINI', dmx_max]
# Get the table index of the parameters above
paper_param_index = []
for pp in paper_params:
# We assume the parameter name are unique in the table
idx = np.where(compare_table2['name'] == pp)[0][0]
paper_param_index.append(idx)
paper_param_index = np.array(paper_param_index)
compare_table2[paper_param_index]
[34]:
| name | Tempo2 Value | T2 unc | units | Tempo2_V-PINT_V | Tempo2_PINT_diff/unct | PINT_unct/Tempo2_unct | no_t_unc |
|---|---|---|---|---|---|---|---|
| str8 | str48 | float128 | object | float128 | float128 | float128 | bool |
| F0 | 277.93771124297462788 | 5.1859268946902080184e-13 | Hz | -6.6613381477509392425e-16 | 0.0012845029023782387781 | 1.0000081311054239701 | False |
| F1 | -7.3387383041227678664e-16 | 4.619148404392432094e-21 | Hz / s | -8.1916189706268323405e-24 | 0.0017734045874857090789 | 0.9999982063295332385 | False |
| FD1 | 3.983282287426775e-05 | 1.6566478062738200598e-06 | s | -1.6361212107284211874e-09 | 0.0009876095598185302606 | 1.00000000325577032 | False |
| FD2 | -1.4729805752137882e-05 | 1.1922596055992699934e-06 | s | 1.4357622807706935195e-09 | 0.0012042362871541143106 | 1.0000000135934370427 | False |
| JUMP1 | -8.7887456483184e-06 | 0.0 | s | -4.9036265484266971897e-11 | 0.00037580263061884339052 | inf | True |
| PX | 0.5061242012322064 | 0.07348886965486496614 | mas | 1.8776131796016670705e-05 | 0.00025549626609032611863 | 1.0000000214259234799 | False |
| ELONG | 244.34767784255382 | 5.95727548431e-09 | deg | 9.1233687271596863866e-12 | 0.0015314666496770878097 | 1.0000013787890671494 | False |
| ELAT | -10.071839047043065 | 3.361025894297e-08 | deg | -1.44861900253090425394e-11 | 0.0004310050109964715927 | 0.99999279185393830885 | False |
| PMELONG | 0.4619096015625491 | 0.010433361011620021289 | mas / yr | 7.4203184182164427796e-06 | 0.0007112107411937685676 | 1.0000025748109051538 | False |
| PMELAT | -7.155145674275822 | 0.058156247552489513664 | mas / yr | -7.1705189014892312116e-05 | 0.0012329748226993851416 | 0.99999266738091230344 | False |
| PB | 14.348465754661366786 | 2.12226632065849e-06 | d | -1.9243153815198810186e-09 | 0.0009067266265257465907 | 0.99999757655418149095 | False |
| A1 | 8.80165312286463 | 8.114047416773300209e-07 | ls | -8.196980871844061767e-10 | 0.0010102209724458007922 | 0.99998413921662954174 | False |
| A1DOT | -4.008979189463729e-15 | 6.2586911221949290846e-16 | ls / s | -1.0335518582171720488e-18 | 0.00165138658872608559 | 1.0000003602893390298 | False |
| ECC | 0.00017372966157521168 | 8.922286680669999241e-09 | 4.158524756102052744e-11 | 0.0046608284455950449096 | 1.0000400945849889922 | False | |
| T0 | 55878.2618994738495070 | 0.00051676746764245482 | d | 4.8831633153723075225e-07 | 0.00094494402630447885357 | 1.0000114310932686899 | False |
| OM | 181.84960401549451478 | 0.01296564244572522874 | deg | 1.225778713571934464e-05 | 0.00094540530382748107187 | 1.0000120115751260204 | False |
| OMDOT | 0.0052395528517645540778 | 0.00135543635075636363 | deg / yr | -1.228657154944533207e-06 | 0.00090646613856778681487 | 0.9999975920019996213 | False |
| M2 | 0.2717633814383356 | 0.08941866471282471085 | solMass | 0.00010426380694084080858 | 0.0011660183841448814885 | 1.0000265499521452384 | False |
| SINI | 0.9064200568225846 | 0.03399283139781983376 | -4.2776490711826653524e-05 | 0.0012583974018289681776 | 1.0000279809291119371 | False | |
| DMX_0010 | 0.00067403356955979 | 0.00020051850482404336064 | pc / cm3 | -3.7725252445390284467e-07 | 0.0018813850860545018075 | 0.99999987947730495375 | False |
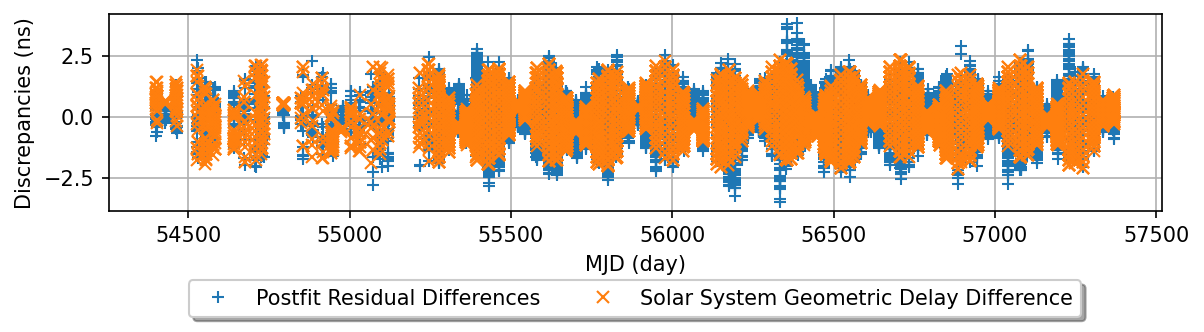
The residual difference between PINT and TEMPO2 is at the level of ~1ns
We believe the discrepancy is mainly from the solar system geometric delay.
We will use the tempo2 postfit parameters, which are wrote out to
J1600-3053_new_tempo2.2.par
[35]:
tempo2_result2 = t2u.general2('J1600-3053_new_tempo2.2.par', tim_file, ['sat', 'pre', 'post', 'freq', 'err'])
m_t22 = models.get_model('J1600-3053_new_tempo2.2.par')
f_t22 = GLSFitter(toas=t, model=m_t22)
f_t22.fit_toas()
tp2_diff_pre2 = f_t22.resids_init.time_resids - tempo2_result2['pre'] * u.s
tp2_diff_post2 = f_t22.resids.time_resids - tempo2_result2['post'] * u.s
WARNING: EPHVER 5 does nothing in PINT [pint.models.timing_model]
WARNING: Unrecognized parfile line 'NE_SW 0' [pint.models.timing_model]
WARNING: Unrecognized parfile line 'NE_SW2 0.000' [pint.models.timing_model]
[36]:
PINT_solar = m_t22.solar_system_geometric_delay(t)
tempo2_solar = t2u.general2('J1600-3053_new_tempo2.2.par', tim_file, ['roemer'])
[37]:
diff_solar = PINT_solar + tempo2_solar['roemer'] * u.s
plt.figure(figsize=(8,2.5), dpi=150)
plt.plot(mjds, (tp2_diff_post2 - tp2_diff_post2.mean()).to_value(u.ns), '+')
plt.plot(mjds, (diff_solar - diff_solar.mean()).to_value(u.ns, equivalencies=[(ls, u.s)]), 'x')
plt.xlabel('MJD (day)')
plt.ylabel('Discrepancies (ns)')
#plt.title('PSR J1600-3053 postfit residual differences between PINT and TEMPO2')
plt.grid(True)
plt.legend(['Postfit Residual Differences', 'Solar System Geometric Delay Difference'],
loc='upper center', bbox_to_anchor=(0.5, -0.3), shadow=True, ncol=2)
plt.tight_layout()
plt.savefig("solar_geo")