This Jupyter notebook can be downloaded from understanding_fitters.ipynb, or viewed as a python script at understanding_fitters.py.
Understanding Fitters
[1]:
from IPython.display import display_markdown
import pint.toa
import pint.models
import pint.fitter
import pint.config
import pint.logging
pint.logging.setup(level="INFO")
[1]:
1
[2]:
%matplotlib inline
import matplotlib.pyplot as plt
# Turn on quantity support for plotting. This is very helpful!
from astropy.visualization import quantity_support
quantity_support()
[2]:
<astropy.visualization.units.quantity_support.<locals>.MplQuantityConverter at 0x7f0e803d2650>
[3]:
# Load some TOAs and a model to fit
m, t = pint.models.get_model_and_toas(
pint.config.examplefile("NGC6440E.par"), pint.config.examplefile("NGC6440E.tim")
)
[4]:
# You can check if a model includes a noise model with correlated errors (e.g. ECORR or TNRED) by checking the has_correlated_errors property
m.has_correlated_errors
[4]:
False
There are several fitters in PINT, each of which is a subclass of Fitter
DownhillWLSFitter- PINT’s workhorse fitter, which does a basic weighted least-squares minimization of the residuals.DownhillGLSFitter- A generalized least squares fitter, like “tempo -G”, that can handle noise processes like ECORR and red noise that are specified by their correlation function properties.WidebandDownhillFitter- A fitter that uses DM estimates associated with each TOA. Also supports generalized least squares.PowellFitter- A very simple example fitter that uses the Powell method implemented in scipy. One notable feature is that it does not require evaluating derivatives w.r.t the model parameters.MCMCFitter- A fitter that does an MCMC fit using the emcee package. This can be very slow, but accomodates Priors on the parameter values and can produce corner plots and other analyses of the posterior distributions of the parameters.WLSFitter,GLSFitter,WidebandFitter- Simpler fitters that make no attempt to ensure convergence.
You can normally use the function pint.fitter.Fitter.auto(toas, model) to construct an appropriate fitter for your model and data.
Weighted Least Squares Fitter
[5]:
# Instantiate a fitter
wlsfit = pint.fitter.DownhillWLSFitter(toas=t, model=m)
A fit is performed by calling fit_toas()
For most fitters, multiple iterations can be limited by setting the maxiter keyword argument.
Downhill fitters will raise the pint.fitter.MaxiterReached exception if they stop before detecting convergence; you can capture this exception and continue if you don’t mind not having the best-fit answer.
[6]:
try:
wlsfit.fit_toas(maxiter=1)
except pint.fitter.MaxiterReached:
print("Fitter has not fully converged.")
Fitter has not fully converged.
[7]:
# A summary of the fit and resulting model parameters can easily be printed
# Only free parameters will have values and uncertainties in the Postfit column
wlsfit.print_summary()
Fitted model using downhill_wls method with 5 free parameters to 62 TOAs
Prefit residuals Wrms = 1090.5801805746107 us, Postfit residuals Wrms = 21.182108789370787 us
Chisq = 59.575 for 56 d.o.f. for reduced Chisq of 1.064
PAR Prefit Postfit Units
=================== ==================== ============================ =====
PSR 1748-2021E 1748-2021E None
EPHEM DE421 DE421 None
CLOCK TT(BIPM2019) TT(BIPM2019) None
UNITS TDB TDB None
START 53478.3 d
FINISH 54187.6 d
TIMEEPH FB90 FB90 None
T2CMETHOD IAU2000B IAU2000B None
DILATEFREQ N None
DMDATA N None
NTOA 0 None
CHI2 59.5747
CHI2R 1.06383
TRES 21.1821 us
POSEPOCH 53750 d
PX 0 mas
RAJ 17h48m52.75s 17h48m52.80034691s +/- 0.00014 hourangle_second
DECJ -20d21m29s -20d21m29.38331083s +/- 0.033 arcsec
PMRA 0 mas / yr
PMDEC 0 mas / yr
F0 61.4855 61.485476554372(18) Hz
PEPOCH 53750 d
F1 -1.181e-15 -1.1813(14)×10⁻¹⁵ Hz / s
TZRMJD 53801.4 d
TZRSITE 1 1 None
TZRFRQ 1949.61 MHz
CORRECT_TROPOSPHERE N None
PLANET_SHAPIRO N None
NE_SW 0 1 / cm3
SWP 2
SWM 0
DM 223.9 224.114(35) pc / cm3
Derived Parameters:
Period = 0.016264003404376±0.000000000000005 s
Pdot = (3.125±0.004)×10⁻¹⁹
Characteristic age = 8.246e+08 yr (braking index = 3)
Surface magnetic field = 2.28e+09 G
Magnetic field at light cylinder = 4806 G
Spindown Edot = 2.868e+33 erg / s (I=1e+45 cm2 g)
[8]:
# The WLS fitter doesn't handle correlated errors
wlsfit.resids.model.has_correlated_errors
[8]:
False
[9]:
# You can request a pretty-printed covariance matrix
cov = wlsfit.get_parameter_covariance_matrix(pretty_print=True)
Parameter covariance matrix:
RAJ DECJ F0 F1 DM
RAJ 1.411e-15
DECJ -2.477e-14 8.328e-11
F0 -5.932e-20 4.079e-17 3.271e-22
F1 1.594e-26 -4.525e-24 -2.080e-29 2.079e-36
DM -6.523e-12 2.067e-08 4.261e-15 2.900e-21 1.221e-03
[10]:
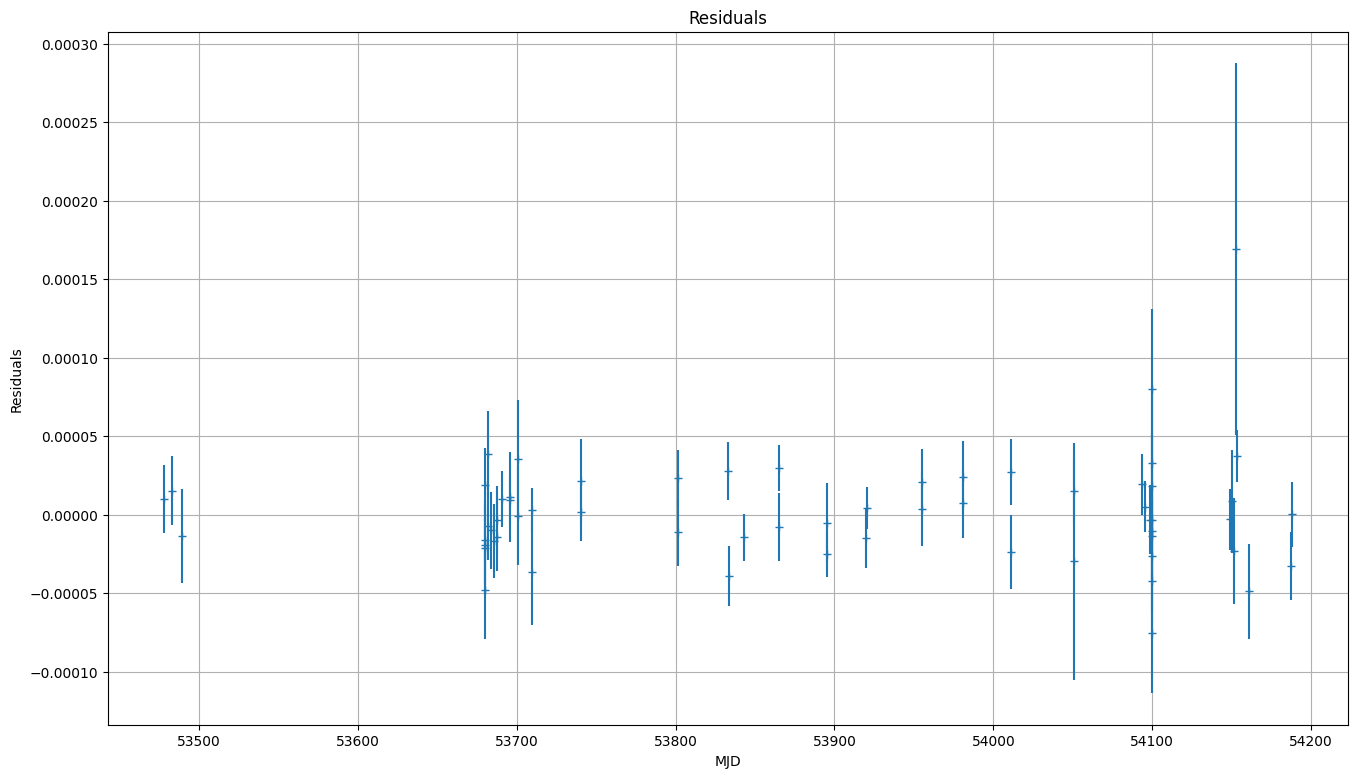
# plot() will make a plot of the post-fit residuals
wlsfit.plot()
Comparing models
There also a convenience function for pretty printing a comparison of two models with the differences measured in sigma.
[11]:
display_markdown(wlsfit.model.compare(wlsfit.model_init, format="markdown"), raw=True)
PARAMETER |
NGC6440E.par |
NGC6440E.par |
Diff_Sigma1 |
Diff_Sigma2 |
|---|---|---|---|---|
PSR |
1748-2021E |
1748-2021E |
||
EPHEM |
DE421 |
DE421 |
||
CLOCK |
TT(BIPM2019) |
TT(BIPM2019) |
||
UNITS |
TDB |
TDB |
||
START |
53478.285871419538264 |
Missing |
||
FINISH |
54187.58732417023191 |
Missing |
||
TIMEEPH |
FB90 |
FB90 |
||
T2CMETHOD |
IAU2000B |
IAU2000B |
||
DILATEFREQ |
False |
False |
||
DMDATA |
False |
False |
||
NTOA |
62 |
0 |
||
CHI2 |
59.574713740962196 |
Missing |
||
CHI2R |
1.0638341739457535 |
Missing |
||
TRES |
21.18210878937078595 |
Missing |
||
POSEPOCH |
53750.0 |
53750.0 |
||
PX |
0.0 |
0.0 |
||
RAJ |
17h48m52.80034691s +/- 0.00014 |
17h48m52.75s +/- 0.05 |
-372.26 |
-1.01 |
DECJ |
-20d21m29.38331083s +/- 0.033 |
-20d21m29s +/- 0.4 |
11.67 |
0.96 |
PMRA |
0.0 |
0.0 |
||
PMDEC |
0.0 |
0.0 |
||
F0 |
61.485476554372(18) |
61.4854765540(5) |
-20.60 |
-0.74 |
PEPOCH |
53750.0 |
53750.0 |
||
F1 |
-1.1813(14)×10⁻¹⁵ |
-1.1810(10)×10⁻¹⁵ |
0.23 |
0.33 |
TZRMJD |
53801.386051200748497 |
53801.386051200748497 |
||
TZRSITE |
1 |
1 |
||
TZRFRQ |
1949.609 |
1949.609 |
||
CORRECT_TROPOSPHERE |
False |
False |
||
PLANET_SHAPIRO |
False |
False |
||
NE_SW |
0.0 |
0.0 |
||
SWP |
2.0 |
2.0 |
||
SWM |
0.0 |
0.0 |
||
DM |
224.114(35) |
223.90(30) |
-6.12 |
-0.71 |
SEPARATION |
0.805131 arcsec |
You can see just how much F1 changed. Let’s compare the \(\chi^2\) values:
[12]:
print(f"Pre-fit chi-squared value: {wlsfit.resids_init.chi2}")
print(f"Post-fit chi-squared value: {wlsfit.resids.chi2}")
Pre-fit chi-squared value: 157920.59715077005
Post-fit chi-squared value: 59.574713740962196
Generalized Least Squares fitter
The GLS fitter is capable of handling correlated noise models.
It has some more complex options using the maxiter, threshold, and full_cov keyword arguments to fit_toas().
If maxiter is less than one, no fitting is done, just the chi-squared computation. In this case, you must provide the residuals argument.
If maxiter is one or more, so fitting is actually done, the chi-squared value returned is only approximately the chi-squared of the improved(?) model. In fact it is the chi-squared of the solution to the linear fitting problem, and the full non-linear model should be evaluated and new residuals produced if an accurate chi-squared is desired.
A first attempt is made to solve the fitting problem by Cholesky decomposition, but if this fails singular value decomposition is used instead. In this case singular values below threshold are removed.
full_cov determines which calculation is used. If True, the full covariance matrix is constructed and the calculation is relatively straightforward but the full covariance matrix may be enormous. If False, an algorithm is used that takes advantage of the structure of the covariance matrix, based on information provided by the noise model. The two algorithms should give the same result up to numerical accuracy where they both can be applied.
To test this fitter properly, we need a model that includes correlated noise components, so we will load one from NANOGrav 9yr data release.
[13]:
m1855 = pint.models.get_model(pint.config.examplefile("B1855+09_NANOGrav_9yv1.gls.par"))
[14]:
# You can check if a model includes a noise model with correlated errors (e.g. ECORR or TNRED) by checking the has_correlated_errors property
m1855.has_correlated_errors
[14]:
True
[15]:
print(m1855)
# Created: 2024-04-18T18:14:09.177154
# PINT_version: 0.9.8+790.g57884b6
# User: docs
# Host: build-24112102-project-85767-nanograv-pint
# OS: Linux-5.19.0-1028-aws-x86_64-with-glibc2.35
# Python: 3.11.6 (main, Feb 1 2024, 16:47:41) [GCC 11.4.0]
# Format: pint
PSR B1855+09
EPHEM DE421
CLK TT(BIPM2019)
UNITS TDB
START 53358.7260000000000000
FINISH 56598.8730000000000000
INFO -f
TIMEEPH FB90
DILATEFREQ N
DMDATA N
NTOA 4005
TRES 5.52
LAMBDA 286.863489330115613 1 0.00000001658590000000
BETA 32.321487755503703 1 0.00000002735260000000
PMLAMBDA -3.2701 1 0.0141
PMBETA -5.0982 1 0.0291
PX 0.2929 1 0.2186
ECL IERS2003
POSEPOCH 54978.0000000000000000
F0 186.4940812707752116 1 3.28468e-11
F1 -6.205147513395e-16 1 1.379566413719e-19
PEPOCH 54978.0000000000000000
CORRECT_TROPOSPHERE N
PLANET_SHAPIRO N
SOLARN0 0.0
SWM 0.0
DM 13.299393
DMX 14.0
DMX_0001 0.015161863 1 0.00351684846
DMXR1_0001 53358.7274600000000001
DMXR2_0001 53358.7784100000000000
DMX_0002 0.0152370685 1 0.00351683449
DMXR1_0002 53420.5489300000000000
DMXR2_0002 53420.5862000000000001
DMX_0003 0.0151895956 1 0.00351649738
DMXR1_0003 53448.4736900000000000
DMXR2_0003 53457.4929400000000000
DMX_0004 0.0151322502 1 0.00351653508
DMXR1_0004 53477.3962800000000000
DMXR2_0004 53477.4345300000000000
DMX_0005 0.0151076504 1 0.00351662711
DMXR1_0005 53532.2328100000000000
DMXR2_0005 53532.2765600000000000
DMX_0006 0.015263814 1 0.00351647013
DMXR1_0006 53603.0363200000000000
DMXR2_0006 53603.0897100000000000
DMX_0007 0.0151897641 1 0.003516599
DMXR1_0007 53628.9652500000000001
DMXR2_0007 53628.9833500000000001
DMX_0008 0.0152890326 1 0.00351651389
DMXR1_0008 53686.7976000000000000
DMXR2_0008 53686.8420300000000001
DMX_0009 0.0152484643 1 0.0035165631
DMXR1_0009 53715.7306400000000001
DMXR2_0009 53715.7667800000000000
DMX_0010 0.0153422398 1 0.00351652136
DMXR1_0010 53750.6227300000000000
DMXR2_0010 53750.6672300000000000
DMX_0011 0.015354092 1 0.00351660466
DMXR1_0011 53798.5010800000000000
DMXR2_0011 53798.5391500000000000
DMX_0012 0.0154295455 1 0.0035165139
DMXR1_0012 53851.3718100000000000
DMXR2_0012 53851.4042800000000000
DMX_0013 0.0154693407 1 0.00351653286
DMXR1_0013 53891.2472700000000000
DMXR2_0013 53891.2851400000000000
DMX_0014 0.0156001615 1 0.00351689266
DMXR1_0014 53926.1632600000000000
DMXR2_0014 53926.1922800000000000
DMX_0015 0.0157477908 1 0.00351642748
DMXR1_0015 53968.0572500000000000
DMXR2_0015 53968.0951700000000000
DMX_0016 0.0159397058 1 0.0035163291
DMXR1_0016 54008.9537200000000000
DMXR2_0016 54008.9864400000000001
DMX_0017 0.0159157339 1 0.00351644531
DMXR1_0017 54043.8368200000000000
DMXR2_0017 54043.8724500000000001
DMX_0018 0.016023921 1 0.00351654202
DMXR1_0018 54092.7126500000000000
DMXR2_0018 54092.7424700000000001
DMX_0019 0.0161119039 1 0.00351660022
DMXR1_0019 54135.5828200000000000
DMXR2_0019 54135.6117100000000000
DMX_0020 0.0163381983 1 0.00351653637
DMXR1_0020 54177.4729700000000000
DMXR2_0020 54177.5136300000000000
DMX_0021 0.0162696012 1 0.00351647068
DMXR1_0021 54472.6627600000000000
DMXR2_0021 54472.6939300000000000
DMX_0022 0.0161627196 1 0.00351654829
DMXR1_0022 54519.5241900000000000
DMXR2_0022 54519.5612300000000000
DMX_0023 0.0162621197 1 0.00351655413
DMXR1_0023 54569.4095400000000000
DMXR2_0023 54569.4667500000000000
DMX_0024 0.0164484026 1 0.00351660433
DMXR1_0024 54819.7148100000000001
DMXR2_0024 54819.7512900000000000
DMX_0025 0.0164104711 1 0.00351663094
DMXR1_0025 54862.5833800000000000
DMXR2_0025 54862.6195900000000001
DMX_0026 0.016569036 1 0.00351659233
DMXR1_0026 54925.4273400000000001
DMXR2_0026 54925.4664099999999999
DMX_0027 0.0167679894 1 0.00351680449
DMXR1_0027 54981.2808400000000000
DMXR2_0027 54981.3303200000000001
DMX_0028 0.0167977392 1 0.00351700298
DMXR1_0028 54998.2094000000000000
DMXR2_0028 54998.2266600000000000
DMX_0029 0.0169021454 1 0.00351669195
DMXR1_0029 55108.9040200000000000
DMXR2_0029 55108.9219600000000000
DMX_0030 0.0170268231 1 0.00351673492
DMXR1_0030 55135.8396000000000000
DMXR2_0030 55135.8607900000000001
DMX_0031 0.0170193727 1 0.0035168257
DMXR1_0031 55170.7389400000000000
DMXR2_0031 55170.7489500000000000
DMX_0032 0.017073879 1 0.0035166332
DMXR1_0032 55205.6495700000000000
DMXR2_0032 55205.6660600000000000
DMX_0033 0.0171315137 1 0.00351779101
DMXR1_0033 55298.4053900000000000
DMXR2_0033 55298.4194000000000001
DMX_0034 0.0172665141 1 0.0035171169
DMXR1_0034 55358.2410600000000000
DMXR2_0034 55358.2606200000000000
DMX_0035 0.0173188687 1 0.00351729708
DMXR1_0035 55391.1726800000000000
DMXR2_0035 55391.1865400000000000
DMX_0036 0.0172177568 1 0.00351711273
DMXR1_0036 55424.0694700000000000
DMXR2_0036 55424.0844000000000000
DMX_0037 0.0172926679 1 0.00351670777
DMXR1_0037 55500.8336900000000000
DMXR2_0037 55500.8483800000000000
DMX_0038 0.0172819014 1 0.00351683726
DMXR1_0038 55528.7626100000000000
DMXR2_0038 55528.7760200000000000
DMX_0039 0.0172309563 1 0.00351654536
DMXR1_0039 55638.4592000000000000
DMXR2_0039 55638.4747300000000000
DMX_0040 0.0172343241 1 0.00351659552
DMXR1_0040 55677.3593500000000000
DMXR2_0040 55677.3764100000000000
DMX_0041 0.0172846259 1 0.00351685651
DMXR1_0041 55701.2862700000000000
DMXR2_0041 55701.3045500000000000
DMX_0042 0.0174295333 1 0.00351754173
DMXR1_0042 55731.2369800000000000
DMXR2_0042 55731.2557500000000000
DMX_0043 0.0174135398 1 0.00351701302
DMXR1_0043 55758.1616300000000000
DMXR2_0043 55758.1828600000000000
DMX_0044 0.0174411084 1 0.00351698583
DMXR1_0044 55789.0676000000000000
DMXR2_0044 55789.0858500000000000
DMX_0045 0.0176716738 1 0.00351656249
DMXR1_0045 55843.9164899999999999
DMXR2_0045 55843.9356300000000001
DMX_0046 0.0178457703 1 0.0035165082
DMXR1_0046 55912.7231500000000000
DMXR2_0046 55912.7409100000000001
DMX_0047 0.0179235129 1 0.00351641019
DMXR1_0047 55989.5168300000000000
DMXR2_0047 55989.5376200000000000
DMX_0048 0.0179551569 1 0.00351643393
DMXR1_0048 56023.4263700000000000
DMXR2_0048 56023.4461900000000000
DMX_0049 0.0180214558 1 0.00351830253
DMXR1_0049 56057.3536400000000000
DMXR2_0049 56057.3537799999999999
DMX_0050 0.0182147041 1 0.00352479233
DMXR1_0050 56106.2205600000000000
DMXR2_0050 56106.2206100000000000
DMX_0051 0.0181602407 1 0.00352162978
DMXR1_0051 56122.1298500000000000
DMXR2_0051 56122.1771700000000000
DMX_0052 0.0180430586 1 0.0035266043
DMXR1_0052 56140.1376700000000000
DMXR2_0052 56140.1377500000000000
DMX_0053 0.0181527986 1 0.00352069768
DMXR1_0053 56166.0571200000000000
DMXR2_0053 56166.0573300000000000
DMX_0054 0.0184943818 1 0.00351633835
DMXR1_0054 56201.9569899999999999
DMXR2_0054 56212.9240400000000000
DMX_0055 0.0182191151 1 0.00351594287
DMXR1_0055 56235.8428200000000000
DMXR2_0055 56235.8681000000000000
DMX_0056 0.0182791281 1 0.00351620206
DMXR1_0056 56254.8200499999999999
DMXR2_0056 56254.8445300000000000
DMX_0057 0.0183448743 1 0.00351757322
DMXR1_0057 56277.7525799999999999
DMXR2_0057 56277.7526300000000000
DMX_0058 0.0182456031 1 0.00351582953
DMXR1_0058 56294.6690800000000000
DMXR2_0058 56306.6485600000000000
DMX_0059 0.0180520318 1 0.00351721613
DMXR1_0059 56319.6395299999999999
DMXR2_0059 56319.6395800000000000
DMX_0060 0.0181223112 1 0.00351582826
DMXR1_0060 56341.5587100000000000
DMXR2_0060 56341.5768099999999999
DMX_0061 0.018162022 1 0.00351580352
DMXR1_0061 56360.5302200000000000
DMXR2_0061 56374.5115599999999999
DMX_0062 0.0181719503 1 0.00351571012
DMXR1_0062 56380.4505300000000000
DMXR2_0062 56380.4684000000000000
DMX_0063 0.0181438391 1 0.00351612171
DMXR1_0063 56411.3969800000000000
DMXR2_0063 56417.3732900000000000
DMX_0064 0.0181367452 1 0.00351601523
DMXR1_0064 56432.3219500000000000
DMXR2_0064 56438.3251700000000000
DMX_0065 0.0181205204 1 0.00351651911
DMXR1_0065 56458.2358500000000000
DMXR2_0065 56458.2537600000000000
DMX_0066 0.0181608231 1 0.00351659645
DMXR1_0066 56479.1780800000000000
DMXR2_0066 56479.1960700000000000
DMX_0067 0.0181467585 1 0.00351643552
DMXR1_0067 56498.1243500000000000
DMXR2_0067 56498.1420100000000000
DMX_0068 0.0182940538 1 0.00351645118
DMXR1_0068 56519.0817400000000000
DMXR2_0068 56519.0981400000000000
DMX_0069 0.0182906491 1 0.00351609445
DMXR1_0069 56538.0092400000000000
DMXR2_0069 56538.0256100000000000
DMX_0070 0.0179611744 1 0.00352468823
DMXR1_0070 56557.9709500000000000
DMXR2_0070 56557.9710100000000000
DMX_0071 0.0183625085 1 0.00351656154
DMXR1_0071 56577.9065599999999999
DMXR2_0071 56577.9228499999999999
DMX_0072 0.0183864669 1 0.003515863
DMXR1_0072 56598.8557200000000000
DMXR2_0072 56598.8720400000000000
BINARY DD
PB 12.32717119132762 1 1.9722e-10
PBDOT 0.0
A1 9.23078048 1 2.03e-07
A1DOT 0.0
E 2.1634e-05 1 2.36e-08
EDOT 0.0
T0 54975.5128660817000000 1 0.0019286695
OM 276.536118059963 1 0.056323656112
OMDOT 0.0
M2 0.233837 1 0.011278
SINI 0.999461 1 0.000178
A0 0.0
B0 0.0
GAMMA 0.0
DR 0.0
DTH 0.0
FD1 0.000161666384 1 3.38650356e-05
FD2 -0.00018821003 1 4.13173074e-05
FD3 0.000107526915 1 2.50177766e-05
T2EFAC -f L-wide_PUPPI 1.507
T2EQUAD -f L-wide_PUPPI 0.25518
T2EFAC -f 430_ASP 1.147
T2EFAC -f L-wide_ASP 1.15
T2EFAC -f 430_PUPPI 1.117
T2EQUAD -f 430_ASP 0.0141
T2EQUAD -f L-wide_ASP 0.42504
T2EQUAD -f 430_PUPPI 0.0264
RNAMP 0.017173
RNIDX -4.91353
TNREDAMP -14.227505410948254
TNREDGAM 4.91353
TNREDC 45.0
ECORR -f 430_PUPPI 0.00601
ECORR -f L-wide_PUPPI 0.31843
ECORR -f L-wide_ASP 0.79618
ECORR -f 430_ASP 0.01117
TZRMJD 54981.2808461648844676
TZRSITE 3
TZRFRQ 424.0
JUMP -fe L-wide -9.449e-06 1 9.439e-06
[16]:
ts1855 = pint.toa.get_TOAs(
pint.config.examplefile("B1855+09_NANOGrav_9yv1.tim"), model=m1855
)
ts1855.print_summary()
Number of TOAs: 4005
Number of commands: 1
Number of observatories: 1 ['arecibo']
MJD span: 53358.727 to 56598.872
Date span: 2004-12-19 17:27:32.961266179 to 2013-11-02 20:55:40.399171358
arecibo TOAs (4005):
Min freq: 422.187 MHz
Max freq: 1760.728 MHz
Min error: 0.05 us
Max error: 17.8 us
Median error: 1.19 us
There is currently a problem with DownhillGLSFitter: it doesn’t record appropriate noise parameters.
[17]:
glsfit = pint.fitter.GLSFitter(toas=ts1855, model=m1855)
[18]:
m1855.DMX_0001.prefix
[18]:
'DMX_'
[19]:
glsfit.fit_toas(maxiter=1)
[19]:
3900.0981532531170273
[20]:
glsfit.print_summary()
Fitted model using generalized_least_square method with 90 free parameters to 4005 TOAs
Prefit residuals Wrms = 5.5386698946489625 us, Postfit residuals Wrms = 1.346783684962562 us
Chisq = 3900.098 for 3914 d.o.f. for reduced Chisq of 0.996
PAR Prefit Postfit Units
=================== ==================== ============================ =====
PSR B1855+09 B1855+09 None
EPHEM DE421 DE421 None
CLOCK TT(BIPM2019) TT(BIPM2019) None
UNITS TDB TDB None
START 53358.7 53358.7 d
FINISH 56598.9 56598.9 d
INFO -f -f None
TIMEEPH FB90 FB90 None
BINARY DD DD None
DILATEFREQ N None
DMDATA N None
NTOA 4005 None
CHI2 3900.1
CHI2R 0.996448
TRES 5.52 1.34678 us
POSEPOCH 54978 d
PX 0.2929 0.30(22) mas
ELONG 286d51m48.56158842s 286d51m48.56156705s +/- 5.9e-05 arcsec
ELAT 32d19m17.35591981s 32d19m17.35582847s +/- 9.8e-05 arcsec
PMELONG -3.2701 -3.270(14) mas / yr
PMELAT -5.0982 -5.099(29) mas / yr
ECL IERS2003 IERS2003 None
F0 186.494 186.49408127078522(27) Hz
PEPOCH 54978 d
F1 -6.20515e-16 -6.20497(25)×10⁻¹⁶ Hz / s
CORRECT_TROPOSPHERE N None
PLANET_SHAPIRO N None
NE_SW 0 1 / cm3
SWP 2
SWM 0
DM 13.2994 pc / cm3
DMX 14 pc / cm3
DMX_0001 0.0151619 0.0152(35) pc / cm3
DMXR1_0001 53358.7 d
DMXR2_0001 53358.8 d
DMX_0002 0.0152371 0.0152(35) pc / cm3
DMXR1_0002 53420.5 d
DMXR2_0002 53420.6 d
DMX_0003 0.0151896 0.0152(35) pc / cm3
DMXR1_0003 53448.5 d
DMXR2_0003 53457.5 d
DMX_0004 0.0151323 0.0151(35) pc / cm3
DMXR1_0004 53477.4 d
DMXR2_0004 53477.4 d
DMX_0005 0.0151077 0.0151(35) pc / cm3
DMXR1_0005 53532.2 d
DMXR2_0005 53532.3 d
DMX_0006 0.0152638 0.0153(35) pc / cm3
DMXR1_0006 53603 d
DMXR2_0006 53603.1 d
DMX_0007 0.0151898 0.0152(35) pc / cm3
DMXR1_0007 53629 d
DMXR2_0007 53629 d
DMX_0008 0.015289 0.0153(35) pc / cm3
DMXR1_0008 53686.8 d
DMXR2_0008 53686.8 d
DMX_0009 0.0152485 0.0153(35) pc / cm3
DMXR1_0009 53715.7 d
DMXR2_0009 53715.8 d
DMX_0010 0.0153422 0.0153(35) pc / cm3
DMXR1_0010 53750.6 d
DMXR2_0010 53750.7 d
DMX_0011 0.0153541 0.0154(35) pc / cm3
DMXR1_0011 53798.5 d
DMXR2_0011 53798.5 d
DMX_0012 0.0154295 0.0154(35) pc / cm3
DMXR1_0012 53851.4 d
DMXR2_0012 53851.4 d
DMX_0013 0.0154693 0.0155(35) pc / cm3
DMXR1_0013 53891.2 d
DMXR2_0013 53891.3 d
DMX_0014 0.0156002 0.0156(35) pc / cm3
DMXR1_0014 53926.2 d
DMXR2_0014 53926.2 d
DMX_0015 0.0157478 0.0158(35) pc / cm3
DMXR1_0015 53968.1 d
DMXR2_0015 53968.1 d
DMX_0016 0.0159397 0.0159(35) pc / cm3
DMXR1_0016 54009 d
DMXR2_0016 54009 d
DMX_0017 0.0159157 0.0159(35) pc / cm3
DMXR1_0017 54043.8 d
DMXR2_0017 54043.9 d
DMX_0018 0.0160239 0.0160(35) pc / cm3
DMXR1_0018 54092.7 d
DMXR2_0018 54092.7 d
DMX_0019 0.0161119 0.0161(35) pc / cm3
DMXR1_0019 54135.6 d
DMXR2_0019 54135.6 d
DMX_0020 0.0163382 0.0163(35) pc / cm3
DMXR1_0020 54177.5 d
DMXR2_0020 54177.5 d
DMX_0021 0.0162696 0.0163(35) pc / cm3
DMXR1_0021 54472.7 d
DMXR2_0021 54472.7 d
DMX_0022 0.0161627 0.0162(35) pc / cm3
DMXR1_0022 54519.5 d
DMXR2_0022 54519.6 d
DMX_0023 0.0162621 0.0163(35) pc / cm3
DMXR1_0023 54569.4 d
DMXR2_0023 54569.5 d
DMX_0024 0.0164484 0.0165(35) pc / cm3
DMXR1_0024 54819.7 d
DMXR2_0024 54819.8 d
DMX_0025 0.0164105 0.0164(35) pc / cm3
DMXR1_0025 54862.6 d
DMXR2_0025 54862.6 d
DMX_0026 0.016569 0.0166(35) pc / cm3
DMXR1_0026 54925.4 d
DMXR2_0026 54925.5 d
DMX_0027 0.016768 0.0168(35) pc / cm3
DMXR1_0027 54981.3 d
DMXR2_0027 54981.3 d
DMX_0028 0.0167977 0.0168(35) pc / cm3
DMXR1_0028 54998.2 d
DMXR2_0028 54998.2 d
DMX_0029 0.0169021 0.0169(35) pc / cm3
DMXR1_0029 55108.9 d
DMXR2_0029 55108.9 d
DMX_0030 0.0170268 0.0170(35) pc / cm3
DMXR1_0030 55135.8 d
DMXR2_0030 55135.9 d
DMX_0031 0.0170194 0.0170(35) pc / cm3
DMXR1_0031 55170.7 d
DMXR2_0031 55170.7 d
DMX_0032 0.0170739 0.0171(35) pc / cm3
DMXR1_0032 55205.6 d
DMXR2_0032 55205.7 d
DMX_0033 0.0171315 0.0171(35) pc / cm3
DMXR1_0033 55298.4 d
DMXR2_0033 55298.4 d
DMX_0034 0.0172665 0.0173(35) pc / cm3
DMXR1_0034 55358.2 d
DMXR2_0034 55358.3 d
DMX_0035 0.0173189 0.0173(35) pc / cm3
DMXR1_0035 55391.2 d
DMXR2_0035 55391.2 d
DMX_0036 0.0172178 0.0172(35) pc / cm3
DMXR1_0036 55424.1 d
DMXR2_0036 55424.1 d
DMX_0037 0.0172927 0.0173(35) pc / cm3
DMXR1_0037 55500.8 d
DMXR2_0037 55500.8 d
DMX_0038 0.0172819 0.0173(35) pc / cm3
DMXR1_0038 55528.8 d
DMXR2_0038 55528.8 d
DMX_0039 0.017231 0.0172(35) pc / cm3
DMXR1_0039 55638.5 d
DMXR2_0039 55638.5 d
DMX_0040 0.0172343 0.0172(35) pc / cm3
DMXR1_0040 55677.4 d
DMXR2_0040 55677.4 d
DMX_0041 0.0172846 0.0173(35) pc / cm3
DMXR1_0041 55701.3 d
DMXR2_0041 55701.3 d
DMX_0042 0.0174295 0.0174(35) pc / cm3
DMXR1_0042 55731.2 d
DMXR2_0042 55731.3 d
DMX_0043 0.0174135 0.0174(35) pc / cm3
DMXR1_0043 55758.2 d
DMXR2_0043 55758.2 d
DMX_0044 0.0174411 0.0174(35) pc / cm3
DMXR1_0044 55789.1 d
DMXR2_0044 55789.1 d
DMX_0045 0.0176717 0.0177(35) pc / cm3
DMXR1_0045 55843.9 d
DMXR2_0045 55843.9 d
DMX_0046 0.0178458 0.0178(35) pc / cm3
DMXR1_0046 55912.7 d
DMXR2_0046 55912.7 d
DMX_0047 0.0179235 0.0179(35) pc / cm3
DMXR1_0047 55989.5 d
DMXR2_0047 55989.5 d
DMX_0048 0.0179552 0.0180(35) pc / cm3
DMXR1_0048 56023.4 d
DMXR2_0048 56023.4 d
DMX_0049 0.0180215 0.0180(35) pc / cm3
DMXR1_0049 56057.4 d
DMXR2_0049 56057.4 d
DMX_0050 0.0182147 0.0182(35) pc / cm3
DMXR1_0050 56106.2 d
DMXR2_0050 56106.2 d
DMX_0051 0.0181602 0.0182(35) pc / cm3
DMXR1_0051 56122.1 d
DMXR2_0051 56122.2 d
DMX_0052 0.0180431 0.0180(35) pc / cm3
DMXR1_0052 56140.1 d
DMXR2_0052 56140.1 d
DMX_0053 0.0181528 0.0182(35) pc / cm3
DMXR1_0053 56166.1 d
DMXR2_0053 56166.1 d
DMX_0054 0.0184944 0.0185(35) pc / cm3
DMXR1_0054 56202 d
DMXR2_0054 56212.9 d
DMX_0055 0.0182191 0.0182(35) pc / cm3
DMXR1_0055 56235.8 d
DMXR2_0055 56235.9 d
DMX_0056 0.0182791 0.0183(35) pc / cm3
DMXR1_0056 56254.8 d
DMXR2_0056 56254.8 d
DMX_0057 0.0183449 0.0183(35) pc / cm3
DMXR1_0057 56277.8 d
DMXR2_0057 56277.8 d
DMX_0058 0.0182456 0.0182(35) pc / cm3
DMXR1_0058 56294.7 d
DMXR2_0058 56306.6 d
DMX_0059 0.018052 0.0181(35) pc / cm3
DMXR1_0059 56319.6 d
DMXR2_0059 56319.6 d
DMX_0060 0.0181223 0.0181(35) pc / cm3
DMXR1_0060 56341.6 d
DMXR2_0060 56341.6 d
DMX_0061 0.018162 0.0182(35) pc / cm3
DMXR1_0061 56360.5 d
DMXR2_0061 56374.5 d
DMX_0062 0.018172 0.0182(35) pc / cm3
DMXR1_0062 56380.5 d
DMXR2_0062 56380.5 d
DMX_0063 0.0181438 0.0181(35) pc / cm3
DMXR1_0063 56411.4 d
DMXR2_0063 56417.4 d
DMX_0064 0.0181367 0.0181(35) pc / cm3
DMXR1_0064 56432.3 d
DMXR2_0064 56438.3 d
DMX_0065 0.0181205 0.0181(35) pc / cm3
DMXR1_0065 56458.2 d
DMXR2_0065 56458.3 d
DMX_0066 0.0181608 0.0182(35) pc / cm3
DMXR1_0066 56479.2 d
DMXR2_0066 56479.2 d
DMX_0067 0.0181468 0.0181(35) pc / cm3
DMXR1_0067 56498.1 d
DMXR2_0067 56498.1 d
DMX_0068 0.0182941 0.0183(35) pc / cm3
DMXR1_0068 56519.1 d
DMXR2_0068 56519.1 d
DMX_0069 0.0182906 0.0183(35) pc / cm3
DMXR1_0069 56538 d
DMXR2_0069 56538 d
DMX_0070 0.0179612 0.0180(35) pc / cm3
DMXR1_0070 56558 d
DMXR2_0070 56558 d
DMX_0071 0.0183625 0.0184(35) pc / cm3
DMXR1_0071 56577.9 d
DMXR2_0071 56577.9 d
DMX_0072 0.0183865 0.0184(35) pc / cm3
DMXR1_0072 56598.9 d
DMXR2_0072 56598.9 d
PB 12.3272 12.32717119133(20) d
PBDOT 0
A1 9.23078 9.23078048(20) ls
A1DOT 0 ls / s
ECC 2.1634e-05 2.1634(24)×10⁻⁵
EDOT 0 1 / s
T0 54975.5 54975.5128(19) d
OM 276.536 276.53(6) deg
OMDOT 0 deg / yr
M2 0.233837 0.234(11) solMass
SINI 0.999461 0.99946(18)
A0 0 s
B0 0 s
GAMMA 0 s
DR 0
DTH 0
FD1 0.000161666 0.000162(34) s
FD2 -0.00018821 -0.00019(4) s
FD3 0.000107527 0.000108(25) s
EFAC1 1.507
EQUAD1 0.25518 us
EFAC2 1.147
EFAC3 1.15
EFAC4 1.117
EQUAD2 0.0141 us
EQUAD3 0.42504 us
EQUAD4 0.0264 us
RNAMP 0.017173
RNIDX -4.91353
TNREDAMP -14.2275
TNREDGAM 4.91353
TNREDC 45
ECORR1 0.00601 us
ECORR2 0.31843 us
ECORR3 0.79618 us
ECORR4 0.01117 us
TZRMJD 54981.3 d
TZRSITE 3 3 None
TZRFRQ 424 MHz
JUMP1 -9.449e-06 -9(9)×10⁻⁶ s
Derived Parameters:
Period = 0.005362100465526423±0.000000000000000008 s
Pdot = (1.78406±0.00007)×10⁻²⁰
Characteristic age = 4.762e+09 yr (braking index = 3)
Surface magnetic field = 3.13e+08 G
Magnetic field at light cylinder = 1.84e+04 G
Spindown Edot = 4.568e+33 erg / s (I=1e+45 cm2 g)
Parallax distance = (3.35±2.45)×10³ pc
Binary model BinaryDD
Mass function = 0.0055573934(4) Msun
Min / Median Companion mass (assuming Mpsr = 1.4 Msun) = 0.2470 / 0.2902 Msun
From SINI in model:
cos(i) = 0.033(5)
i = 88.12(31) deg
Pulsar mass (Shapiro Delay) = 1.2804782086531232 solMass
The GLS fitter produces two types of residuals, the normal residuals to the deterministic model and those from the noise model.
[21]:
glsfit.resids.time_resids
[21]:
[22]:
glsfit.resids.noise_resids
[22]:
{'pl_red_noise': <Quantity [-1.27090740e-06, -1.27090740e-06, -1.27090740e-06, ...,
-1.27096267e-06, -1.27096267e-06, -1.27096267e-06] s>,
'ecorr_noise': <Quantity [-7.68053637e-11, -7.68053637e-11, -7.68053637e-11, ...,
3.75218951e-07, 3.75218951e-07, 3.75218951e-07] s>}
[23]:
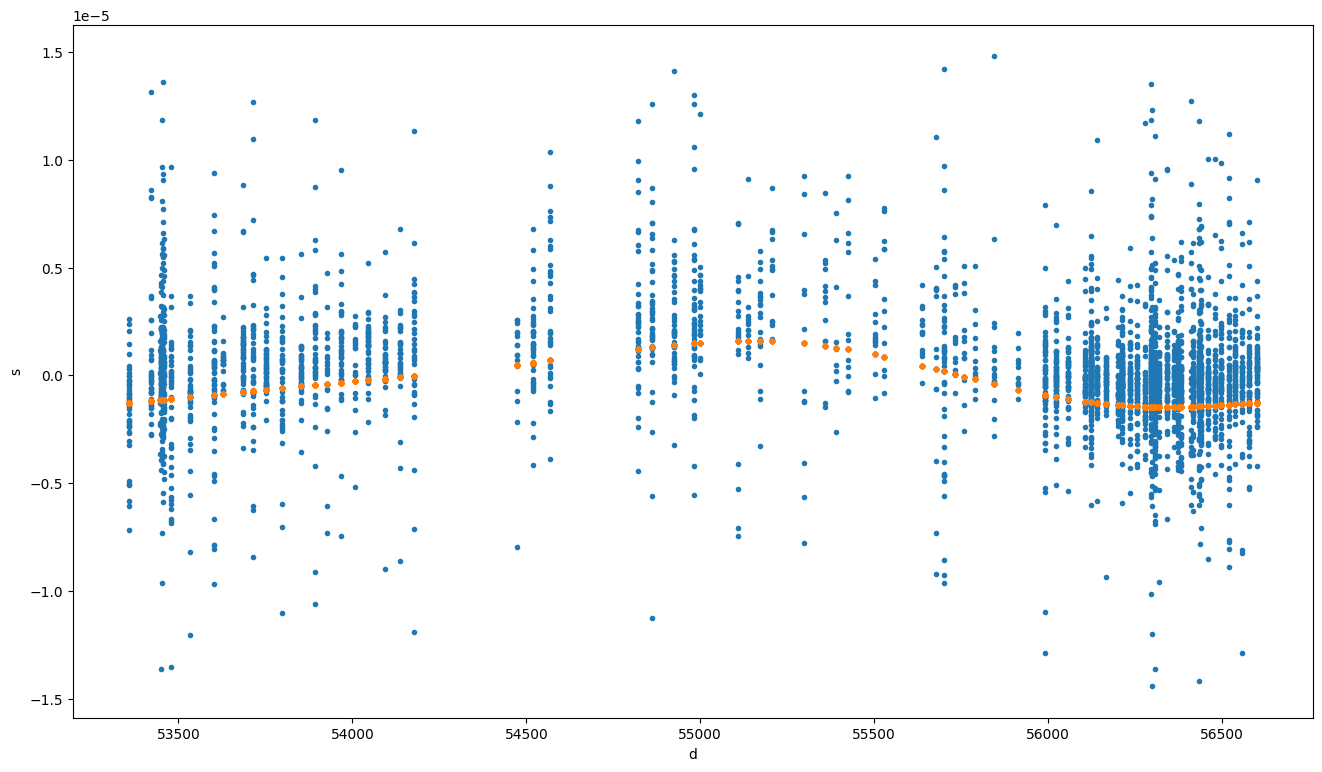
# Here we can plot both the residuals to the deterministic model as well as the realization of the noise model residuals
# The difference will be the "whitened" residuals
fig, ax = plt.subplots(figsize=(16, 9))
mjds = glsfit.toas.get_mjds()
ax.plot(mjds, glsfit.resids.time_resids, ".")
ax.plot(mjds, glsfit.resids.noise_resids["pl_red_noise"], ".")
[23]:
[<matplotlib.lines.Line2D at 0x7f0e79c04350>]
Choosing fitters
You can use the automatic fitter selection to help you choose between WLSFitter, GLSFitter, and their wideband variants. The default Downhill fitters generally have better performance than the plain variants.
[24]:
autofit = pint.fitter.Fitter.auto(toas=ts1855, model=m1855)
[25]:
autofit.fit_toas()
[25]:
True
[26]:
display_markdown(autofit.model.compare(glsfit.model, format="markdown"), raw=True)
PARAMETER |
B1855+09_NANOGrav_9yv1.gls.par |
B1855+09_NANOGrav_9yv1.gls.par |
Diff_Sigma1 |
Diff_Sigma2 |
|---|---|---|---|---|
PSR |
B1855+09 |
B1855+09 |
||
EPHEM |
DE421 |
DE421 |
||
CLOCK |
TT(BIPM2019) |
TT(BIPM2019) |
||
UNITS |
TDB |
TDB |
||
START |
53358.727464829469664 |
53358.727464829469664 |
||
FINISH |
56598.871995360779607 |
56598.871995360779607 |
||
INFO |
-f |
-f |
||
TIMEEPH |
FB90 |
FB90 |
||
BINARY |
DD |
DD |
||
DILATEFREQ |
False |
False |
||
DMDATA |
False |
False |
||
NTOA |
4005 |
4005 |
||
CHI2 |
3900.098975108194 |
3900.098153253117 |
||
CHI2R |
0.9964483840337746 |
0.9964481740554719 |
||
TRES |
1.3465976125409702851 |
1.3467836849625620117 |
||
POSEPOCH |
54978.0 |
54978.0 |
||
PX |
0.30(22) |
0.30(22) |
0.00 |
0.00 |
ELONG |
286d51m48.56156715s +/- 5.9e-05 |
286d51m48.56156705s +/- 5.9e-05 |
-0.00 |
-0.00 |
ELAT |
32d19m17.35582891s +/- 9.7e-05 |
32d19m17.35582847s +/- 9.8e-05 |
-0.00 |
-0.00 |
PMELONG |
-3.270(14) |
-3.270(14) |
0.00 |
0.00 |
PMELAT |
-5.099(29) |
-5.099(29) |
-0.00 |
-0.00 |
ECL |
IERS2003 |
IERS2003 |
||
F0 |
186.49408127078522(27) |
186.49408127078522(27) |
0.00 |
0.00 |
PEPOCH |
54978.0 |
54978.0 |
||
F1 |
-6.20497(25)×10⁻¹⁶ |
-6.20497(25)×10⁻¹⁶ |
-0.00 |
-0.00 |
CORRECT_TROPOSPHERE |
False |
False |
||
PLANET_SHAPIRO |
False |
False |
||
NE_SW |
0.0 |
0.0 |
||
SWP |
2.0 |
2.0 |
||
SWM |
0.0 |
0.0 |
||
DM |
13.299393 |
13.299393 |
||
PB |
12.32717119133(20) |
12.32717119133(20) |
-0.00 |
-0.00 |
PBDOT |
0.0 |
0.0 |
||
A1 |
9.23078048(20) |
9.23078048(20) |
-0.00 |
-0.00 |
A1DOT |
0.0 |
0.0 |
||
ECC |
2.1634(24)×10⁻⁵ |
2.1634(24)×10⁻⁵ |
0.00 |
0.00 |
EDOT |
0.0 |
0.0 |
||
T0 |
54975.5129(8) |
54975.5128(19) |
-0.08 |
-0.03 |
OM |
276.536(23) |
276.53(6) |
-0.08 |
-0.03 |
OMDOT |
0.0 |
0.0 |
||
M2 |
0.234(11) |
0.234(11) |
0.00 |
0.00 |
SINI |
0.99946(18) |
0.99946(18) |
-0.00 |
-0.00 |
A0 |
0.0 |
0.0 |
||
B0 |
0.0 |
0.0 |
||
GAMMA |
0.0 |
0.0 |
||
DR |
0.0 |
0.0 |
||
DTH |
0.0 |
0.0 |
||
FD1 |
0.000162(34) |
0.000162(34) |
0.00 |
0.00 |
FD2 |
-0.00019(4) |
-0.00019(4) |
-0.00 |
-0.00 |
FD3 |
0.000108(25) |
0.000108(25) |
0.00 |
0.00 |
EFAC1 |
1.507 |
1.507 |
||
EQUAD1 |
0.25518 |
0.25518 |
||
EFAC2 |
1.147 |
1.147 |
||
EFAC3 |
1.15 |
1.15 |
||
EFAC4 |
1.117 |
1.117 |
||
EQUAD2 |
0.0141 |
0.0141 |
||
EQUAD3 |
0.42504 |
0.42504 |
||
EQUAD4 |
0.0264 |
0.0264 |
||
RNAMP |
0.017173 |
0.017173 |
||
RNIDX |
-4.91353 |
-4.91353 |
||
TNREDAMP |
-14.227505410948254 |
-14.227505410948254 |
||
TNREDGAM |
4.91353 |
4.91353 |
||
TNREDC |
45.0 |
45.0 |
||
ECORR1 |
0.00601 |
0.00601 |
||
ECORR2 |
0.31843 |
0.31843 |
||
ECORR3 |
0.79618 |
0.79618 |
||
ECORR4 |
0.01117 |
0.01117 |
||
TZRMJD |
54981.28084616488447 |
54981.28084616488447 |
||
TZRSITE |
3 |
3 |
||
TZRFRQ |
424.0 |
424.0 |
||
JUMP1 |
-9(9)×10⁻⁶ |
-9(9)×10⁻⁶ |
0.00 |
0.00 |
SEPARATION |
0.000000 arcsec |
The results are (thankfully) identical.
The MCMC fitter is considerably more complicated, so it has its own dedicated walkthroughs in MCMC_walkthrough.ipynb (for photon data) and examples/fit_NGC6440E_MCMC.py (for fitting TOAs).